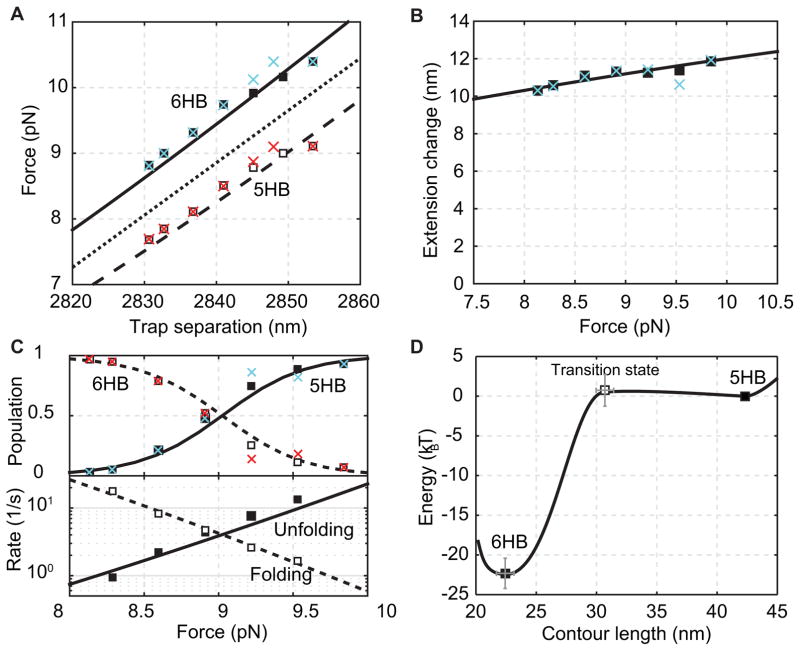
Fig. 7.
Comparison of properties of the gp41 hairpin transition derived from the histogram analysis (crosses), the HMM analysis (squares) and best model fitting (dashed and solid curves). (A) Average forces of the folded state (solid square or cyan cross) and the unfolded state (hollow square or red cross) as a function of trap separation. The mean force is shown as the dotted line. (B) Extension difference between the folded and unfolded states as a function of the mean force. (C) State populations (top) and transition rates (bottom) as a function of the mean force. (D) Simplified energy landscape of gp41 hairpin folding at zero force determined by the model fitting in A–C.