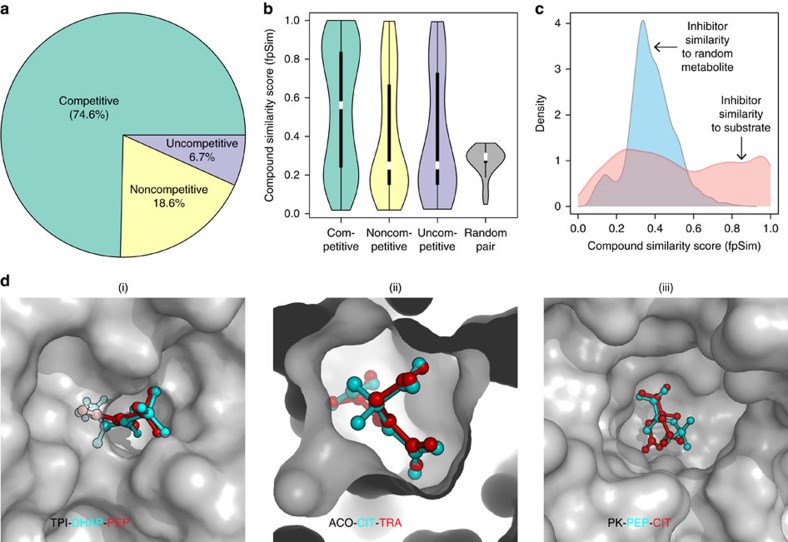
Figure 2. Enzyme inhibition across the metabolic landscape is driven by structural similarity between metabolites.
(a) Gold-standard set: Competitive inhibition is the most common type of inhibition (74.6%) followed by noncompetitive (18.6%) and then by uncompetitive inhibition (6.7%), across 462 examples determined in individual enzymological experiments (Supplementary Data 1). (b) Pairwise compound similarity (fpSim) between inhibitor and substrate reveals significant structural similarity (0=non-similar, 1=maximum possible structural similarity). The median similarity for allosteric inhibitors is not significantly different to that of a random inhibitor; however, the spread is much higher, with a about 1/3rd of allosteric inhibitors being as similar enzyme’s metabolic substrates as competitive inhibitors. (c) The genome-scale set of enzymatic inhibitors have a wide range of similarities including highly similar metabolite–inhibitor relationships (ranging from 0.2–1), compared to random substrates, which are not structurally similar (0.2–0.4). (d) Three selected examples (protein/metabolite structure determined by X-ray crystallography) revealing the extent of structural similarity between competitive inhibitor and substrate in the active site of key enzymes. (i) Triosephosphate isomerase (TPI: 1NEY, inhibitor: 4OWG (ref. 5)); Substrate: Dihydroxyacetone phosphate (DHAP); Inhibitor: phosphoenolpyruvate (PEP), (ii) Enzyme: Aconitase (ACO: 1C96 (ref. 28), inhibitor: 1ACO (ref. 29)); Substrate: Citrate (CIT); Inhibitor: Trans-aconitate (TRA), (iii) Enzyme: Pyruvate Kinase (PK: 4HYV (ref. 26), inhibitor: 4IP7 (ref. 27)); Substrate: phosphoenolpyruvate (PEP); Inhibitor: Citrate (CIT).