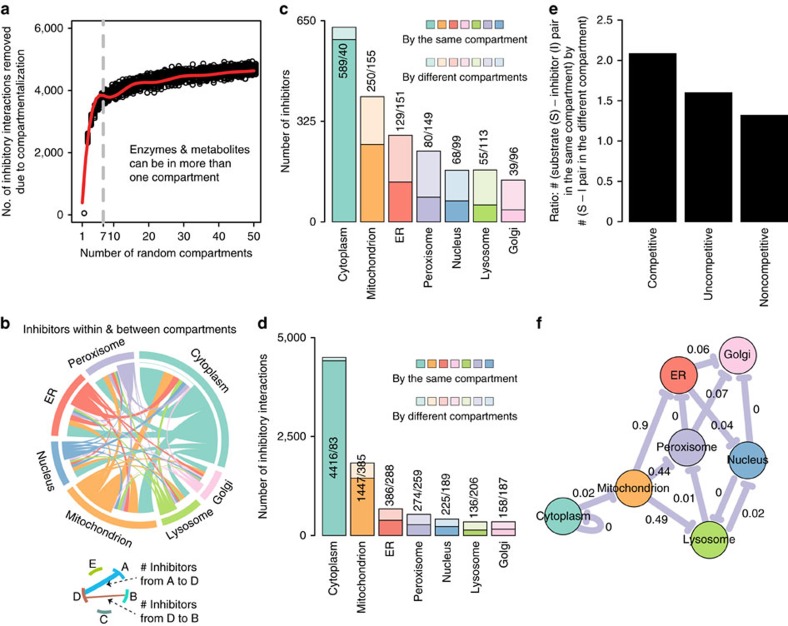
Figure 5. Compartmentalization considerably reduces metabolic self-inhibition.
(a) Computational simulation by randomly placing inhibitors and enzymes in random compartments, maintaining a metabolic network organization. red line: bs spline fitting; grey line: seven organellar compartments as in the humans. (b) Human network: compartmentalization globally reduces inhibitor–enzyme interactions. (c,d) Compartmentalization protects organellar metabolic reactions from enzyme inhibition (in c expressed as the number of inhibitors, in d as the number of inhibitory interactions). (e) Compartmentalization is more effective in preventing allosteric inhibition, as on preventing competitive inhibition. (f) Compartment–compartment specific inhibition network in which nodes represent the seven subcellular compartments. Edges are drawn if the number of inhibitor compounds between a pair of compartments is significantly enriched (P value<0.05). FDR values are highlighted over edges. Metabolic self-inhibition is enriched in the cytoplasm as expected (Figs 1), but not within the organelles. Instead, organellar metabolites would significantly enrich inhibition if localized to other organelles (P value<0.05).