Summary
Distinct signaling pathways activate the NF-κB family of transcription factors. The canonical NF-κB-signaling pathway is mediated by IκB kinase 2/β (IKK2/β), while the non-canonical pathway depends on IKK1/α. The structural and biochemical bases for distinct signaling by these otherwise highly similar IKKs are unclear. We report single-particle cryoelectron microscopy (cryo-EM) and X-ray crystal structures of human IKK1 in dimeric (∼150 kDa) and hexameric (∼450 kDa) forms. The hexamer, which is the representative form in the crystal but comprises only ∼2% of the particles in solution by cryo-EM, is atrimer of IKK1 dimers. While IKK1 hexamers are not detectable in cells, the surface that supports hexamer formation is critical for IKK1-dependent cellular processing of p100 to p52, the hallmark of non-canonical NF-κB signaling. Comparison of this surface to that in IKK2 indicates significant divergence, and it suggests a fundamental role for this surface in signaling by these kinases through distinct pathways.
Graphical abstract
The non-canonical NF-κB-signaling pathway, essential for development, activates processing of NF-κB2/β100 into p52. Two protein kinases, NIK and IKK1/α, are central components of this pathway. Polley et al. identify a unique surface on IKK1/α based on the X-ray and cryo-EM structures, and they find that it is essential for NIK interaction and p100 processing.

Introduction
NF-κB family transcription factors are central to cellular immune and inflammatory responses and cell survival in higher eukaryotes (Baldwin, 2001; Xiao and Ghosh, 2005). Loss of regulation and consequent aberrant NF-κB activity lead to various pathological conditions, including chronic inflammatory and metabolic diseases, autoimmune disorders, and cancer (Ben-Neriah and Karin, 2011; Grivennikov et al., 2010; Xia et al., 2014). Under homeostatic conditions, NF-κB activity is maintained at a low, basal level through cytoplasmic sequestration via noncovalent binding to IκB (inhibitor of NF-κB) proteins (Hinz and Scheidereit, 2014). The transcriptional potential of NF-κB is realized upon IκB removal as a result of activation of one of two distinct NF-κB-signaling pathways, referred to as canonical and non-canonical (also known as alternative) (Bonizzi et al., 2004). Two highly homologous Ser/Thr kinases, IKK2/IKKβ and IKK1/IKKα, act as central regulators of these two pathways (Ghosh and Karin, 2002; Sun, 2012).
IKK2, which is present within a holo-IKK complex that is presumed to contain also IKK1 and the NEMO/IKKγ scaffolding protein, regulates the canonical pathway of NF-κB activation that is essential for inflammation and innate immunity in response to pathogens (DiDonato et al., 1997). IKK2 is primarily responsible (Li et al., 1999) for the phosphorylation of specific residues of prototypical IκB proteins (IκBα, -β, and -ε) bound to NF-κB as well as an atypical IκB protein, NF-κB1/p105, which is the precursor of the NF-κB subunit p50 (Karin and Ben-Neriah, 2000). Phosphorylation-dependent ubiquitylation and proteasomal degradation of the IκB proteins (or partial processing of p105 to p50) lead to the release of active NF-κB dimers (Hayden and Ghosh, 2008).
In contrast, IKK1 regulates NF-κB signaling through the non-canonical pathway, which is essential for lymphoid organogenesis and adaptive immunity. IKK1 phosphorylates specific residues of NF-κB2/β100 on its C-terminal IκBδ segment. This leads to processing of p100 and generation of the NF-κB subunit p52 (Claudio et al., 2002; Coope et al., 2002; Dejardin et al., 2002; Senftleben et al., 2001; Sun, 2012). Interestingly, generation of p52 through the non-canonical pathway is critically dependent on another kinase domain-containing cellular factor, NF-κB-inducing kinase (NIK) (Xiao et al., 2001, 2004), but it does not rely on either IKK2 or NEMO. In resting cells, NIK level is kept low through its continuous proteasome-dependent degradation (Qing et al., 2005; Vallabhapurapu et al., 2008; Zarnegar et al., 2008). However, upon stimulation of cells by non-canonical ligands, and in certain malignant cells, NIK is stabilized from proteolysis to the extent that it becomes capable of recruiting and inducing the kinase activity of IKK1. Kinase activities of both NIK and IKK1 are essential for efficient processing of p100 into p52 (Sun, 2012). IKK1 and NIK phosphorylate three serines (866, 870, and 872) of NF-κB2/β100 within its C-terminal IκBδ segment, leading to the processing of p100 to p52.
The X-ray crystal structure of Xenopus IKK2 provided insight into domain organization, dimer assembly, and substrate-binding features in IKK family kinases (Xu et al., 2011). Subsequent X-ray structures of human IKK2 provided further insights into the mechanism through which IKK2 becomes catalytically active (Polley et al., 2013). However, these IKK2 structures have failed to shed light on the question of how either IKK1 or IKK2, in response to different upstream stimuli, act to regulate such a wide range of NF-κB-dependent activities (Hinz and Scheidereit, 2014; Scheidereit, 2006). As part of our effort to understand the mechanistic basis underlying the unique signaling function of IKK1, we report the three-dimensional (3D) structures of human IKK1 determined by combining cryoelectron microscopy (cryo-EM) and X-ray crystallography. These structures reveal an unexpected trimeric arrangement of IKK1 dimers that gives rise to a hexamer. Cellular and biochemical studies indicate that the IKK1 hexameric assembly is of insufficient stability to be clearly observed in cell. However, mutation of residues on IKK1 that mediate its hexamerization results in a drastic impairment of the non-canonical NF-κB-signaling pathway. This study provides a glimpse into the structure of IKK1, reveals how structural features that are unique to IKK1 contribute to its role in non-canonical NF-κB signaling, and identifies possible tools and approaches for the specific regulation of signal-induced activity of IKK1.
Results
Determination of Human IKK1 Structure by Cryo-EM and X-Ray Crystallography
In an effort to crystallize human IKK1 (hIKK1), we generated various truncation mutants with both wild-type (WT) and the S176E,180E (EE) double mutant in which two activation loop serine residues were replaced with phosphomimetic glutamates (Figure 1A). Only one construct of hIKK1 (residues 10–667; EE) reproducibly yielded crystals suitable for X-ray diffraction analysis in the presence of the Calbiochem IKK inhibitor XII (Christopher et al., 2007). However, attempts at phase determination by molecular replacement, using IKK2 dimers in various conformations (Liu et al., 2013; Polley et al., 2013; Xu et al., 2011) as search models, failed. This was somewhat unexpected, as sequence alignments and homology modeling analyses predicted the domain arrangement and mode of dimerization for IKK1 to be similar to those in IKK2. The two proteins share ∼50% identity and 70% homology (Figure S1).
Figure 1. Determination of the Structure of Human IKK1.

(A) Domain organization is as follows: KD, kinase domain; ULD, ubiquitin-like domain; SDD, scaffold dimerization domain; and NBD, NEMO-binding domain. Functionally relevant residues and those mutated in this study are marked.
(B) Representative 2D class averages of cryo-EM particles of hIKK1 are shown.
(C and D) Initial cryo-EM model (C) and refined cryo-EM map (D) of the hIKK1 dimer without classification are shown.
(E) Crystallographic model of an hIKK1 dimer derived from the X-ray data is shown.
(F) hIKK1 protomer with its three domains in different shades of red is shown.
To prepare better search models for molecular replacement, we turned to single-particle cryo-EM. The expected species of IKK1 in solution is a dimer with a molecular weight (MW) of ∼150 kDa, which is considered small by cryo-EM standards. However, various recent advances have pushed the resolution limit and reduced the size limitation of the approach (Bai et al., 2015; Cheng, 2015). Micrographs of purified IKK1 revealed particles that resembled IKK2 dimers and yielded an initial map with clearly resolved domains and a-helical features (Figures 1B-1D; Figures S2A and S2B). This cryo-EM map was used to dock individual domains of an IKK1 homology model that was generated using a crystal structure of IKK2 (PDB: 4KIK). The resultant model yielded a molecular replacement solution for our X-ray diffraction data (Table S1). Further analyses of the cryo-EM data enabled us to refine structural models to ∼5.5 Å (discussed later).
Features of Human IKK1 Structure
The cryo-EM and X-ray crystal structures revealed that, like other IKK family members IKK2 and TBK1, IKK1 forms a dimer (Figure 1E) and the individual IKK1 subunit is composed of three ordered protein domains: a kinase domain (KD), a ubiquitin-like domain (ULD), and a scaffold dimerization domain (SDD) (Figure 1F). The backbone architectures of individual domains are highly conserved among IKK family proteins.
The overall structural features of IKK1 observed in cryo-EM and X-ray maps were highly consistent; however, we used the X-ray map to describe the structure due to better resolution. The KD adopts the familiar fold observed in other canonical protein kinases. Backbone electron density for the activation loop is clearly visible (Figure S2C), and positioning of important catalytic site residues indicates that the KD is in an active conformation. Density for the inhibitor is relatively well defined, and we were able to position it sensibly within the Fo-Fc omit density map (Figure S2D). The ULD, which has been implicated previously in substrate specificity (Gray et al., 2014), indeed adopts a fold that is similar to that of ubiquitin (Figure 1F). It also contains a unique ∼17-residue loop insertion spanning residues 367–383, the general path of which is visible in the density. The SDD, a long stalklike structure composed of an anti-parallel three-helix bundle, provides an extensive surface for dimerization on one end. Two long α helices, α2 s and α6 s of the SDD, run parallel to each other and are accompanied by a discontinuous, anti-parallel third helical segment composed of α3 s, α4 s, and α5 s (Figure 1F). The dimer interfaces of IKK1 and IKK2 are highly similar, containing conserved residues and burying ∼3,400-Å2 surface area (Figure S1) (Polley et al., 2013; Xu et al., 2011).
Despite global similarity among IKK monomers, their tertiary organizations reflect significant differences in relative placement of all three domains. The most pronounced difference between IKK1 and IKK2 was observed in the orientation of the KD relative to the SDD and its stably associated ULD. Previous studies indicated that spatial arrangement and inter-domain connectivity are critical for the activity of IKK2. In IKK2, the KD is positioned appropriately with respect to the SDD and ULD through interactions of several residues in ULD (L389 and F390), SDD (W434 and H435), and KD (F111 and E112). Mutation of these residues reduced kinase activity of IKK2, with W434 and H435 mutants displaying a drastic reduction (Polley et al., 2013). Interestingly, most of these residues are conserved in IKK1, except for W434 and F111 (Figure S1).
A major advantage of single-particle cryo-EM is the ability to detect structural heterogeneity within the data. To assess the conformational plasticity of IKK1, we analyzed particles using maximum likelihood-based 3D classification and simultaneous refinement (Lyumkis et al., 2013). This analysis revealed three predominant conformational states of IKK1 dimers (with an approximately uniform distribution), the maps of which were subsequently refined to average global resolutions of ∼5.6, 5.4, and 5.2 Å (Figures S3A–S3D; Table S2). The resulting maps enabled independent construction of three molecular models describing IKK1 flexibility using recently developed protocols implemented within Rosetta and Phenix (Figure 2A). These models display differing angles between apposing KDs and suggest mobility of IKK1 domains (Figure S3E). Interestingly, the splaying of two KDs in all six independent dimers within the X-ray lattice is similar (Figure S3F), the extent of which is similar to the least-splayed dimer model from cryo-EM (Figure 2B; Figure S3G). Overall, our X-ray and cryo-EM analyses revealed distinct conformers of the IKK1 dimer that are reminiscent of the conformational plasticity observed in IKK2 (Polley et al., 2013) and could explain the difficulty in obtaining well-diffracting IKK1 crystals.
Figure 2. Cryo-EM and X-Ray Structures of Human IKK1.
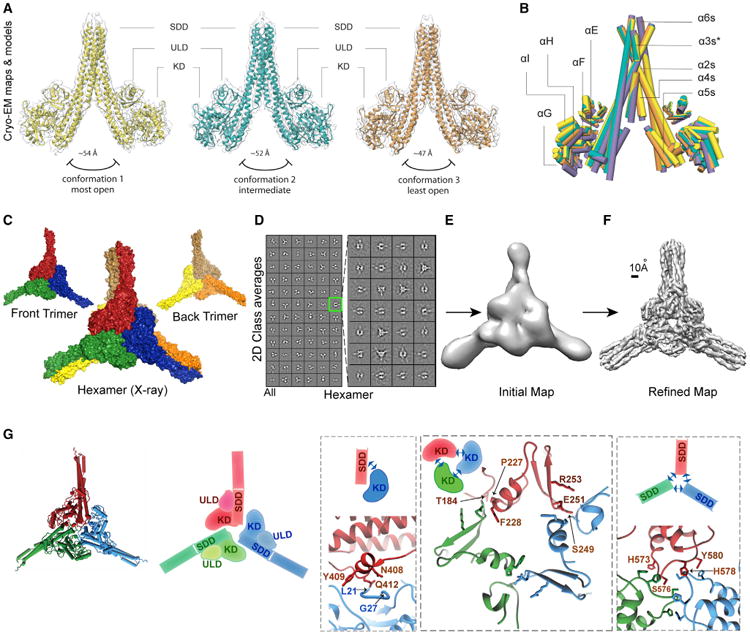
(A) Atomic models of three different conformers superimposed on a transparent isosurface representation of their respective cryo-EM maps are shown.
(B) Superposition of three IKK1 dimer conformers modeled from the cryo-EM maps (orange, yellow, and green) and the X-ray model (purple) is shown as pipes.
(C) Crystallographic model of the IKK1 hexamer (surface representation) with constituent trimers is shown.
(D) Left panel: representative 2D cryo-EM class averages of hIKK1 representing the complete dataset (this panel is identical to Figure 1B). Right panel: 2D class averages of hIKK1 after sub-classification of the class outlined in green are shown.
(E) Initial cryo-EM map of IKK1 hexamer calculated from sub-classified class averages is shown.
(F) Cryo-EM map of IKK1 hexamer refined from raw particles to ∼5.9-Å resolution is shown.
(G) A cartoon and a schematic representation of the trimer interface formed by monomers from three dimers constituting the hexamer. Zoomed-in views showthe three interactive patches (KD:SDD, KD:KD, and SDD:SDD) of the interface with interacting residues highlighted.
IKK1 Self-Assembles into a Trimer of Dimers, a Hexamer
The most intriguing finding observed in both X-ray and cryo-EM data is the assembly of three IKK1 dimers into a hexamer. The asymmetric unit contained two independent hexamers, each of which formed through the self-association of three IKK1 dimers (Figure 2C). We also detected a small population of particles (∼2%) in the cryo-EM data that did not conform to particles yielding high-resolution dimer structures (Figure 2D). A careful inspection of these 1,432 particles through iterative two-dimensional (2D) sub-classification (Figure 2D, right panel) resulted in a density map of ∼5.9-Å resolution that contained a D3-symmetric IKK1 hexamer (Figures 2E and 2F; Figures S3I–S3K). It is likely that the 3-fold symmetry of the hexamer was selected for the single dimer conformation observed within the crystal; this observation was supported by a more uniform local resolution for the reconstructed EM map of the hexamer (Figure S3I).
The observation of hexameric IKK1 assemblies by two independent methods and the overall similarity between crystallographic and EM hexamers suggest that hexamerization through a trimer interface is specific to and an inherent property of the IKK1 protein. Moreover, since cryo-EM analyses were performed under solution conditions, in the absence of any inhibitor, and at low protein concentration, assembly of IKK1 into hexamers is likely to have biological relevance.
Three IKK1 dimers associate with 3-fold symmetry to form the IKK1 hexamer. The close-knit trimer interface involves the KD and SDD of each protomer and buries a surface area of about 6,300 A2. Interacting patches of the buried trimer interface can be classified into three groups: KD-KD, KD-SDD, and SDD-SDD (Figure 2G). In the KD-SDD interface, residues within the L21-G27 stretch from the KD of one protomer are in close proximity to N408, Y409, and Q412 of the SDD in another protomer. Residues within the H573-Y580 loop of SDD from all three protomers closely associate to form the SDD-SDD interface. In the KD-KD interface, residues in the S249-E251 loop closely appose P227-F228. Interestingly, despite their high degree of sequence and structural homology, most of these interacting residues at the trimer interface in IKK1 are not conserved in IKK2, providing an explanation for the absence of a similar assembly in IKK2.
Overexpressed IKK1 Exists as Dimers in the Cell
The specific interaction of IKK1 dimers at the trimer interface prompted our investigation as to whether IKK1 exists as a hexamer in cells and whether perturbation of residues within the trimer interface destabilizes hexamerization and non-canonical NF-κB signaling. Within the trimer interface, we selected residues for mutation that could potentially disrupt inter-protomer interactions without perturbing kinase catalytic fold and/or activity. We concentrated primarily on residues of the SDD to disrupt both SDD-SDD and SDD-KD interactions. A double-mutant IKK1HY (H578A Y580A, SDD-SDD) was chosen for this assay. IKK1WT and IKK1HY with native activation loop sequences were overexpressed as HA-fusion proteins in 293T cells, and their oligomerization status was monitored by size-exclusion chromatography of S100 extracts followed by immunoblot analysis of the fractions with anti-HA antibody (Figure 3A). Both overexpressed IKK1WT and IKK1HY were found to be mostly dimeric.
Figure 3. Oligomerization Property of IKK1 in Cells.

(A) Size-exclusion profiles of overexpressed IKK1 (WT and HY mutant) and IKK2 in S100 extracts of 293T cells. Protein levels are assessed by western blot and normalized band intensities are plotted.
(B) Size-exclusion profiles of IKK1 (1–676) and IKK2 (1–700) proteins expressed in insect cells and purified to homogeneity. Absorbance at 280 nm reflects the level of proteins.
(C) Size-exclusion profiles of endogenous IKK1 in S100 extract of MEF cells and HA-IKK1WTand HA-IKK1HYin S100 extract of stably reconstituted ikk1−/− MEF cells. Protein levels are assessed by western blot.
Analysis of insect cell-expressed pure IKK1 by Superose 6 size-exclusion chromatography also revealed the dimer to be the predominant form of IKK1 in solution, with only a very small fraction eluting as a large oligomer (Figure 3B). Similar size-exclusion experiments aimed at assessing the oligomeric state of purified IKK2 indicated that it exists exclusively as a dimer (Figure 3B), and IKK2 in mammalian cell extracts is known to exist predominantly as a dimer. Overall, these results clearly show that overexpressed IKK1 exists predominantly as a dimer. It is noteworthy that native IKK1 in mouse embryonic fibroblast (MEF) 3T3 cells predominantly migrates within an uncharacterized large complex, as indicated by a western blot analysis for IKK1 (Figure 3C), likely in association with IKK2 and NEMO (∼1 MDa). We reconstituted ikk1−/− MEF cell lines with HA-IKK1 WT and HA-IKK1HY, and we compared the oligomeric properties of WT and mutant IKK1 proteins, where expression levels of the proteins are in a similar range to that of the native protein. These experiments revealed that the mutant behaved similarly to the WT protein. However, these fractionation experiments were not sensitive enough to clearly distinguish the presence of a minor population of oligomeric IKK1 from dimeric free IKK1 or the uncharacterized holo-IKK complex.
Perturbation of the IKK1 Trimer Interface Leads to Pathway-Specific Inhibition of NF-κB Activation
It has been established that formation of a NEMO:IKK2 complex is indispensable for the activation of NF-κB through the canonical pathway. In the non-canonical pathway, however, the interaction between NIK and IKK1 is essential. To clarify the distinct role of IKK1 in the non-canonical signaling, we assayed for IKK-specific signaling events in MEF cells. Consistent with previous findings, we observed that processing of p100 to p52 upon treatment with lymphotoxin β receptor agonist antibody (α-LTβR) is drastically reduced in ikk1−/− MEF cells compared to that in WT and ikk2−/− MEF cells (Figure S4A). In contrast, canonical NF-κB signaling, as reflected by rapid degradation of lκBα substrate in response to TNF-α, is severely compromised only in ikk2−/− MEF cells and not in WT or ikk1−/− MEFs (Figure S4B). These differences in cellular functions also were observed in an in vitro kinase assay where IKK1 and IKK2 preferentially phosphorylated the IκBδ portion of p100 and lκBα, respectively (Figure S4C). These observations underscore the functional uniqueness of two otherwise highly homologous kinases IKK1 and IKK2, which share ∼70% homology, an identical domain arrangement, and a nearly identical kinase activation segment.
We wished to investigate if the structural surfaces that mediate the unique trimerization of IKK1 dimers into hexamers might be critical for non-canonical signaling in vivo. We tested IKK1HY (H578A Y580A) and another double mutant, IKK1NY (N408A Y409A), that target the SDD-KD patch of the trimer interface. Processing of p100 to p52 was monitored in ikk1−/− MEF cells retrovirally transduced with IKK1HY and IKK1NY in response to α-LTbR treatment over different time periods. These reconstituted MEFs failed to exhibit any appreciable processing of p100 to p52, while the extent and kinetics of p100 processing in ikk1−/− MEF cells reconstituted with native IKK1 (IKK1WT) were similar to that of WT MEFs (Figure 4A). This reflects a severe disruption of NF-κB activation through the non-canonical pathway, with IKK1 harboring trimer interface mutations. In both IKK1HY and IKK1NY reconstituted cells, the characteristic lκBα degradation profile upon treatment with TNF-α was observed (Figure 4B), confirming that the canonical NF-κB-signaling pathway is mostly unaffected. These findings suggest that trimer interface interactions are essential for non-canonical NF-κB signaling but have little or no apparent relevance in canonical signaling.
Figure 4. Mutant IKK1 Abrogates Non-canonical NF-κB Signaling.

(A)ikk1−/− MEF cells fail to induce p100 processing in response to α-LTbR when trimer interface mutants of IKK1 (IKK1HYand IKK1NY) are introduced. Introduction of native IKK1 enables processing similarly to the WT MEF; signaling is monitored by immunoblotting with anti-p52.
(B) IKK1 trimer interface mutants do not affect the characteristic lκBα degradation profile in response to TNF-α treatment during canonical NF-κB signaling, as monitored by immunoblotting with anti-lκBα.
(C) The trimer interface mutant (IKK1HY) shows a defect in binding to NIK when compared to WT IKK1. NIK recruitment efficiencies of native and mutant IKK1 proteins were assessed in a co-immunoprecipitation assay.
(D) Residues of IKK1 involved in forming the trimer interface and those of IKK2 in tetramer-forming interfaces in the crystal lattice are shown in context of the respective monomers. Overlapping patches in SDD and KD that contain interacting residues are highlighted. A few prominent residues and the general active site area are marked.
Since propagation of a non-canonical signal depends on productive association between IKK1 and NIK, we next tested whether the surface of IKK1 containing HY is required for the recruitment of IKK1 to NIK. Different amounts of HA-tagged IKK1 (WT and HY) were mixed with constant amounts of Myc-NIK, all proteins being expressed in HEK293T cells and obtained from its total extract. Immunoprecipitation with anti-Myc antibody and western blot analysis with anti-HA antibody indicated that the trimer interface-defective (IKK1HY) mutant exhibited partial reduction in binding to NIK as compared to the WT IKK1 (Figure 4C).
Discussion
In this study, we identified a surface on IKK1 that is essential for the processing of p100 to p52 through the non-canonical NF-κB-signaling pathway. Initial experiments suggest that this surface is involved in the interaction of IKK1 with NIK. It is possible that the IKK1:NIK interaction through this surface is essential for IKK1 activation, which is one of the key initial events in the propagation of non-canonical signaling. Intriguingly, residues from the homologous surface on IKK2 have been shown to be essential for its activation (Figure 4D). We speculate that the two IκB kinases may undergo activation by similar mechanisms. In the case of IKK2, the surface in question might be responsible for a signal-dependent interaction with NEMO. This signal-dependent interaction is separate from the stable binding observed between NEMO and IKK2 through their N and C termini, respectively.
Intriguingly, these functionally important homologous surfaces of IKK1 and IKK2 were both identified on account of their involvement in self-assembly within their respective crystal lattices (also from the solution structure of IKK1). We have thus far failed to relate self-assembly of IKK1 (or IKK2) to their function in cells. It is still possible that aberrant activation upon unintended interaction with NIK or other non-canonical signaling factor(s) is curbed through dynamic engagement of this surface through self-oligomerization in unstimulated cells. We also cannot rule out the possibility that regulated formation of IKK1 oligomers leads to the appropriate docking of partner proteins and thereby its activation. It is known that, only after NIK is permitted to accumulate within the cell and become activated via non-canonical signaling, the proper activation of IKK1 can occur through its interaction with NIK and p100. Thus, insight into the dynamic interplay of these three factors is key to understanding non-canonical NF-κB signaling. Our study highlights important surface patches in IKK1 that could likely be targeted to prevent its assembly with NIK and p100 and, thus, selectively block the activation of NF-κB through the non-canonical pathway.
One of the critical unresolved questions regarding IKK is the composition of IKK complex(es) in vivo. The predominant form of IKK is presumed to be a heterotrimeric IKK complex (IKK1: IKK2:NEMO). This complex, which appears to be in the high-MW range (∼1 MDa), as judged by size-exclusion chromatography, has been studied extensively by different groups since its discovery (Li et al., 2001; Miller and Zandi, 2001; Solt et al., 2009). It is possible that NEMO, which interacts with both IKK1 and IKK2, is responsible for the elution of the complex as high MW. The canonical signaling triggered by stimuli, such as TNF-α and lipopolysaccharide (LPS), activates this high-MW complex, as indicated by lκBα phosphorylation activity of IKK2 contained within this pool. It is unclear whether IKK1 present in this complex propagates a non-canonical NF-κB signal. Our structures and preliminary cellular signaling data set up a platform to further investigate the biochemical nature and forms of IKK1 that could be targeted by NIK during non-canonical signaling.
Experimental Procedures
Protein expression, purification, X-ray structure determination, cryo-EM data acquisition, cryo-EM image analysis, IKK1-dimer model building and refinement from cryo-EM data, immunoprecipitation assays, in vitro kinase assays (IP-KA), retroviral reconstitution of MEF cells, cellular signaling assays, and size-exclusion chromatography to assess oligomeric status are described in detail in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The X-ray and EM structures reveal that IKK1/α assembles as a trimer of dimers
Mutation of the trimer-forming surface abrogates processing of NF-κB2/β100 into p52
The trimer-forming surface is important for interaction with NIK
Acknowledgments
The full-length human IKK1 (hIKK1) cDNA clone and α-LTbR and a-p52 antibodies were provided by Drs. M. Karin, C. Ware, and N. Rice, respectively. This study was supported by NIH grants AI064326, CA141722, and GM071862 to G.G.; NIH grant AI048034; Cancer Center Core grant (P30 CA014195-38); the H.N. and Frances C. Berger Foundation to I.M.V.; the Leona M. and Harry B. Helmsley Charitable Trust grant 2012-PGMED002 to I.M.V.; and NIH DP5 OD021396-01 to D.L. We thank Drs. J. Lazarz, A. Joachimiak, and S. Ginell (SBC-19ID) and Drs. R. Rajashankar and D. Neau (NECAT-24ID) at Advanced Photon Source (APS) for help with crystal data collection; B. Anderson and J.C. Ducom at The Scripps Research Institute for help with EM data collection and network infrastructure; and J. Fitzpatrick and F. Dwyer for computational support at Salk. Molecular graphics and analyses were performed with the University of California, San Francisco (UCSF) Chimera (supported by National Institute of General Medical Sciences [NIGMS] P41-GM103311) and/or the PyMol package. The authors thank Drs. A. Hoffmann, T. Huxford, S. Basak, and T. Hunter for discussions and T. Huxford for editing the manuscript. I.M.V. is an American Cancer Society Professor. S.P. is presently a Wellcome Trust DBT India Alliance Intermediate Fellow.
Footnotes
Accession Numbers: The accession numbers for the EM maps of the three conformers of IKK1 dimer and that for the IKK1 hexamer reported in this paper are EMDataBank: EMD-8436, EMD-8437, EMD-8438, and EMD-8439, respectively. The accession numbers for the coordinates for the three conformers of IKK1 dimer refined using EM maps reported in this paper are PDB: 5TQW, 5TQX, and 5TQY. The accession number for the atomic coordinates and structure factors of human IKK1 model refined from crystallographic data reported in this paper is PDB: 5EBZ.
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.10.067.
Author Contributions: S.P., D.-B.H., T.B., D.L., and G.G. designed experiments. S.P. designed the protein constructs and optimized protein expression, purification, and crystallization. S.P., D.-B.H., and T.B. collected and processed X-ray data. D.-B.H. obtained the first molecular replacement (MR) solution and S.P., D.-B.H., and T.B. refined the model from X-ray data. S.P. and D.-B.H. analyzed the X-ray structure. D.O.P. and D.L. collected and processed cryo-EM data. D.L. built and refined atomic models from cryo-EM data. S.P., M.C.M., A.M., and T.B. performed functional assays. I.M.V. provided direction, guidance, helpful feedback on analysis, and support. S.P., T.B., D.L., and G.G. wrote the manuscript, and all authors contributed to the writing of manuscript.
References
- Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Baldwin AS., Jr Series introduction: the transcription factor NF-kappaB and human disease. J Clin Invest. 2001;107:3–6. doi: 10.1172/JCI11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- Bonizzi G, Bebien M, Otero DC, Johnson-Vroom KE, Cao Y, Vu D, Jegga AG, Aronow BJ, Ghosh G, Rickert RC, Karin M. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher JA, Avitabile BG, Bamborough P, Champigny AC, Cutler GJ, Dyos SL, Grace KG, Kerns JK, Kitson JD, Mellor GW, et al. The discovery of 2-amino-3,5-diarylbenzamide inhibitors of IKK-alpha and IKK-beta kinases. Bioorg Med Chem Lett. 2007;17:3972–3977. doi: 10.1016/j.bmcl.2007.04.088. [DOI] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Gray CM, Remouchamps C, McCorkell KA, Solt LA, Dejardin E, Orange JS, May MJ. Noncanonical NF-κB signaling is limited by classical NF-κB activity. Sci Signal. 2014;7:ra13. doi: 10.1126/scisignal.2004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hinz M, Scheidereit C. The IκB kinase complex in NF-κB regulation and beyond. EMBO Rep. 2014;15:46–61. doi: 10.1002/embr.201337983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, Fang X, Gaynor RB. Role of IKKgamma/nemo in assembly of the Ikappa B kinase complex. J Biol Chem. 2001;276:4494–4500. doi: 10.1074/jbc.M008353200. [DOI] [PubMed] [Google Scholar]
- Liu S, Misquitta YR, Olland A, Johnson MA, Kelleher KS, Kriz R, Lin LL, Stahl M, Mosyak L. Crystal structure of a human IκB kinase b asymmetric dimer. J Biol Chem. 2013;288:22758–22767. doi: 10.1074/jbc.M113.482596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BS, Zandi E. Complete reconstitution of human IkappaB kinase (IKK) complex in yeast. Assessment of its stoichiometry and the role of IKKgamma on the complex activity in the absence of stimulation. J Biol Chem. 2001;276:36320–36326. doi: 10.1074/jbc.M104051200. [DOI] [PubMed] [Google Scholar]
- Polley S, Huang DB, Hauenstein AV, Fusco AJ, Zhong X, Vu D, Schröfelbauer B, Kim Y, Hoffmann A, Verma IM, et al. A structural basis for IκB kinase 2 activation via oligomerization-dependent trans autophosphorylation. PLoS Biol. 2013;11:e1001581. doi: 10.1371/journal.pbio.1001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Qu Z, Xiao G. Stabilization of basally translated NF-kappaB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-kappaB2 p100. J Biol Chem. 2005;280:40578–40582. doi: 10.1074/jbc.M508776200. [DOI] [PubMed] [Google Scholar]
- Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, Greten FR, Krähn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Solt LA, Madge LA, May MJ. NEMO-binding domains of both IKKalpha and IKKbeta regulate IkappaB kinase complex assembly and classical NF-kappaB activation. J Biol Chem. 2009;284:27596–27608. doi: 10.1074/jbc.M109.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. The noncanonical NF-κB pathway. Immunol Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Ghosh S. NF-kappaB, an evolutionarily conserved mediator of immune and inflammatory responses. Adv Exp Med Biol. 2005;560:41–45. doi: 10.1007/0-387-24180-9_5. [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- Xu G, Lo YC, Li Q, Napolitano G, Wu X, Jiang X, Dreano M, Karin M, Wu H. Crystal structure of inhibitor of kB kinase β. Nature. 2011;472:325–330. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


