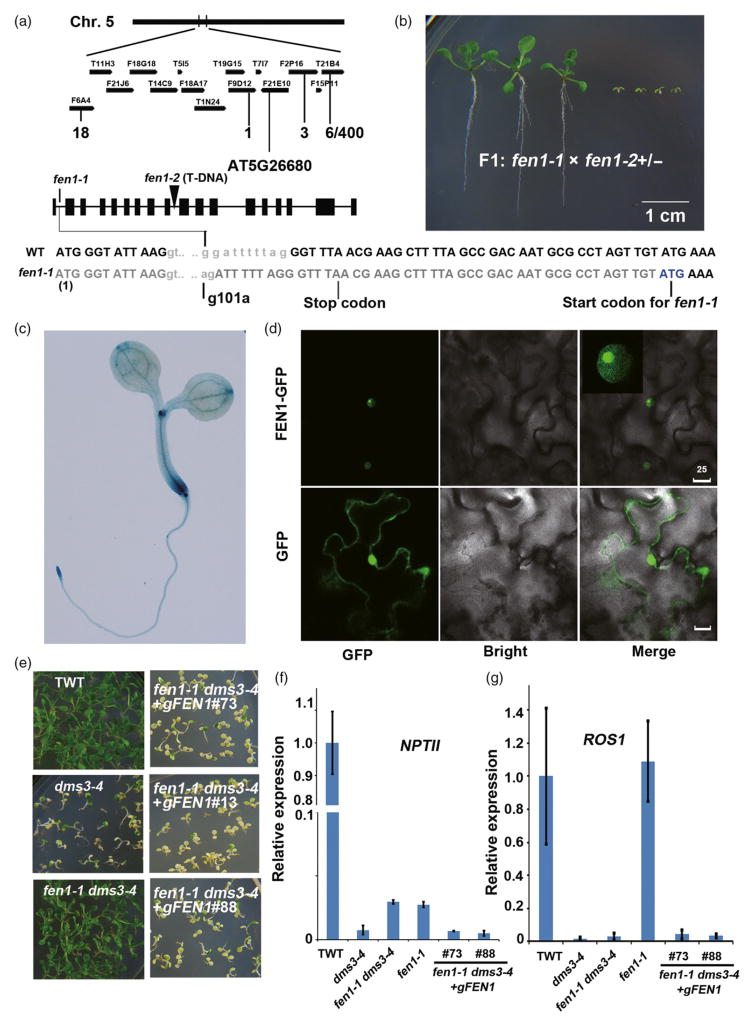
Figure 1.
Map-based cloning of FEN1 and complementation assay.(a) Map-based cloning of FEN1. The mutation was localized on chromosome 5 (Chr. 5) in the region between the F9D12 and F2P16 bacterial artificial chromosomes (BACs). The recombination rates were indicated under the matching BACs. A G to A mutation was identified 101 bp from ATG at gene At5 g26680. The transcript of the mis-splicing caused by the mutation led to the formation of a stop codon. An ATG downstream of the stop codon produced by the mutation might be used as a translation start site for the fen1-1 mutant. Therefore, fen1-1 produced a protein lacking 17 amino acids at the N-terminus. (b) Genetic analysis of the different fen1 alleles. Phenotype of the F1 progeny of fen1-1 crossed with fen1-2(+/−) heterozygotes. (c) Expression pattern of FEN1. GUS staining of the ProFEN1:GUS transgenic plant. (d) Subcellular localization of FEN1-GFP. FEN1 fused with GFP was transiently expressed in tobacco leaf cells. The 35S-GFP construct was used as a control. Bar = 25 μm. (e) fen1-1 complementation. Phenotype of the transgenic wild type (TWT), dms3-4, fen1-1 dms3-4 and three independent genomic DNA complementation lines (#76, #13 and #88) in the fen1-1 dms3-4 background on MS supplied with 50 mg L−1 kanamycin. (f) Relative expression of NPTII among TWT, dms3-4, fen1-1 dms3-4 and two independent genomic DNA complementation lines. (g) Relative expression of ROS1 among TWT, dms3-4, fen1-1 dms3-4 and two independent genomic DNA complementation lines.