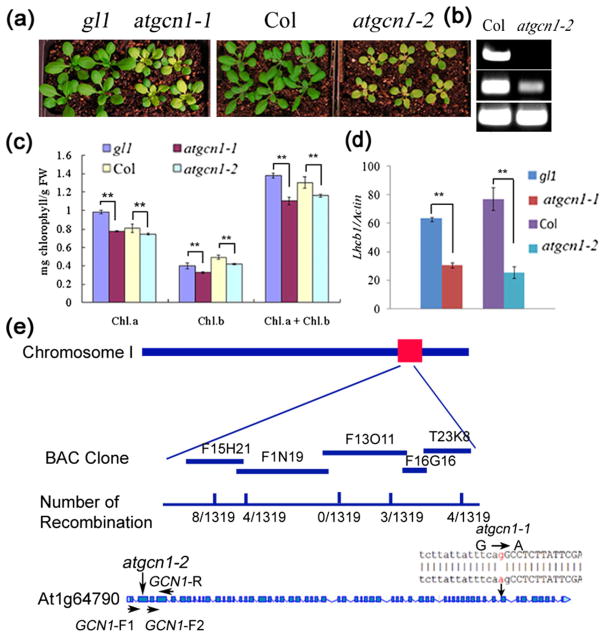
Figure 1.
Map-based cloning of AtGCN1. (a) The new leaves of 4-week-old seedlings of atgcn1-1 and atgcn1-2 grown in soil are yellow. (b) Transcription of AtGCN1 in atgcn1-2. PCR products were amplified by primers across the T-DNA insertion site of atgcn1-2 (AtGCN1-F1 and AtGCN1-R) (top) and downstream of the T-DNA insertion site of atgcn1-2 (AtGCN1-F2 and AtGCN1-R) (middle). Actin was used as the control (bottom). (c) Chl content in atgcn1-1 and atgcn1-2. A t-test was performed between atgcn1-1, atgcn1-2, and the wild type. Double asterisks indicate p <0.01. (d) Quantitative analysis of the expression of LHCB1 in atgcn1-1 and atgcn1-2 grown in soil. Double asterisks indicate p <0.01. (f) Map-based cloning of AtGCN1. The mutation was mapped to the bottom of chromosome I. Fine mapping was performed in the BAC clones F15H21, F1N19, F13O11, F16G16 and T23K8, and the numbers of recombinations at the corresponding BAC clones are listed. A mutation from G to A was identified in the 3'-splicing site of the 52nd intron of AT1G64790.1. In atgcn1-2 (SALK_063702C), a T-DNA was inserted in the second exon of AT1G64790.1.