FIGURE 6.
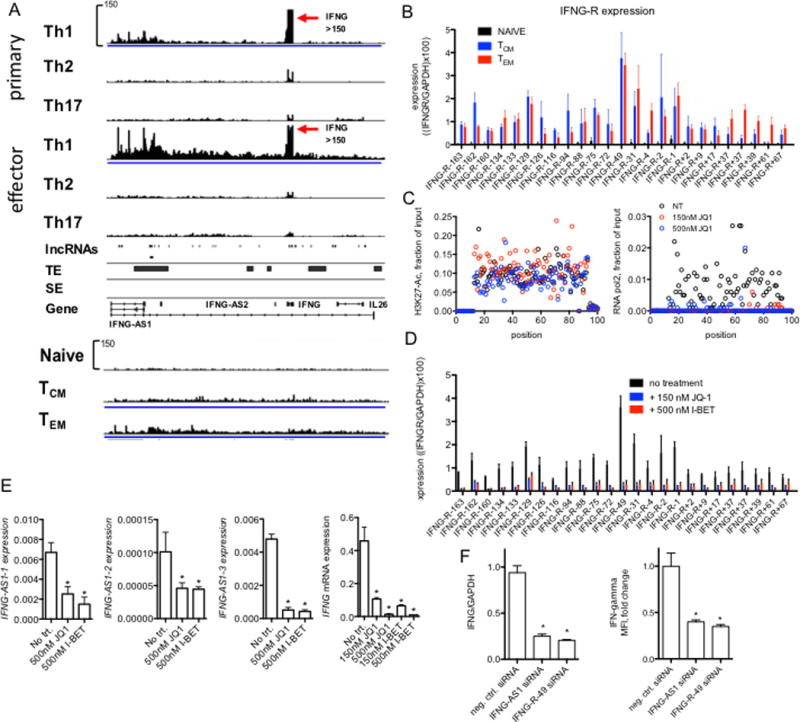
Novel lncRNAs at the IFNG locus. (A) RNA-seq tracks spanning ~300 kb from IFNG-AS1 past IL26 in primary and effector Th1, Th2 and Th17 cells, naïve CD4+ T cells and TCM and TEM cells. Below are positions of genomic loci encoding novel lncRNAs determined by de novo assembly, genomic positions of typical and super enhancers (TE & SE, respectively) identified in total CD4+ TM cells are also shown (from ref. 20). (B) PCR validation of RNA-seq results normalized to GAPDH. Each novel lncRNA is named relative to the IFNG transcriptional start site, - indicates toward the p-terminus, + toward the q-terminus, differences between either TCM and TN or TEM and TN were statistically significant, N=4, P < 0.05. (C) JQ1 treatment of memory T cells abrogates RNA polymerase II binding but not H3K27Ac across the IFNG locus. Indicated concentrations of JQ1 were added to memory T cells cultures. Cells were stimulated with plate-bound anti-CD3 for 24 hours and levels of H3K27-Ac (left panel) and RNA polymerase II (right panel) were determined by ChIP. For this analysis, we designed a tiling array. 100 PCR primer pairs were designed at ~3 kb intervals to span the 300-kb IFNG locus. Data are expressed as mean fraction of input (n=3) P < 0.05, ANOVA. (D) Total memory CD4+ T cells were cultured for 24 hr with JQ1 or I-BET to displace BRD-containing proteins from genomic loci. RNA was purified and levels of the indicated IFNG locus novel lncRNAs, IFNG-R-#, determined by PCR. Results are expressed relative to levels of GAPDH. P < 0.05, treated versus untreated (N=4). (E) JQ1 or I-BET inhibit IFNG-AS1 and IFNG expression. Total memory CD4+ T cells were treated with and without JQ1 or I-BET for 24 hr and stimulated with anti-CD3 for an additional 24 hr. Levels of three unique isoforms of IFNG-AS1 designated 1, 2, 3 (Fig. 6A), and IFNG were determined by PCR and normalized to levels of GAPDH, *=P < 0.05 (N=4). (F) Individual siRNAs targeting IFNG-AS1 or IFNG-R-49 were transfected into total memory CD4+ T cells prior to stimulation with anti-CD3 for 24 hours. Transfected cells were isolated by fluorescence activated cell sorting prior to mRNA analysis or to measure intracellular levels of IFN-g. P values were determined by unpaired T-test. Error bars are mean +/− S.D. * = p < 0.05.
