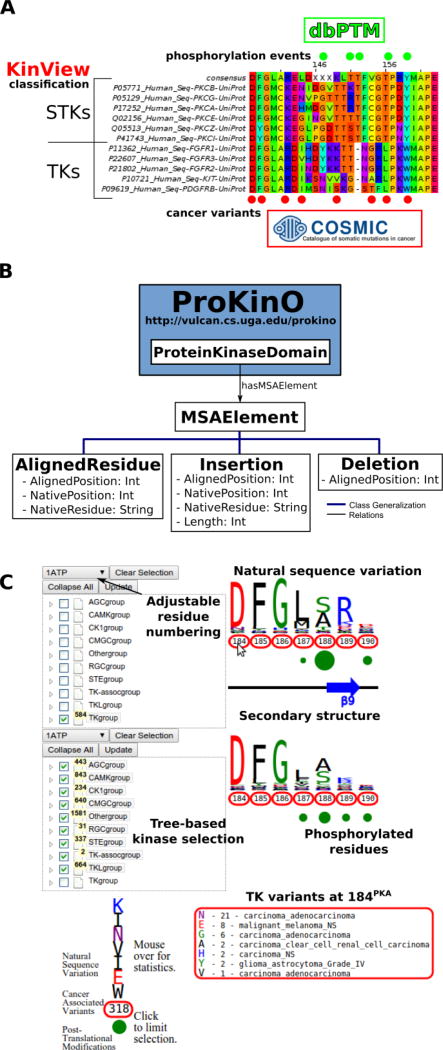
Figure 1.
A) Example benefits of integrative analysis. Placing cancer variants in the context of natural sequence variation and post-translational modifications provides mechanistic insight as to how cancer variants can disrupt functionally significant residues, like phosphorylation sites, to deregulate kinase catalytic activity and rewire signaling networks. Using the classification of the human kinome allows us to consider differences between the kinase groups, families and subfamilies. B) MSAOnt schema. MSAOnt consists of three main classes: AlignedResidue, Insertion and Deletion. Using these three classes, we can fully represent an alignment of sequences to a profile or consensus. It is connected to the ProKinO ProteinKinaseDomain class through the hasMSAElement relation. C) The KinView interface. The interface is divided horizontally into top and bottom regions. Each region has an associated tree structure to select the kinase group, family, subfamily or domains of interest. The pulldown menu above the tree adjusts the residue numbering to match any Human kinase UniProt numbering. After clicking ‘Update’, the natural sequence variation of the selected kinases is displayed using a weblogo. Red circumscribed residue numbers show positions with cancer associated variants, while green circles show positions with experimentally validated post-translational modifications (PTMs). The secondary structure is displayed between the top and bottom regions. Detailed information is displayed by hovering the mouse over a residue number (cancer variants) or green circle (PTMs).