Figure 2. TGIRT enzyme delivers higher signal and lower background for DMS-MaPseq.
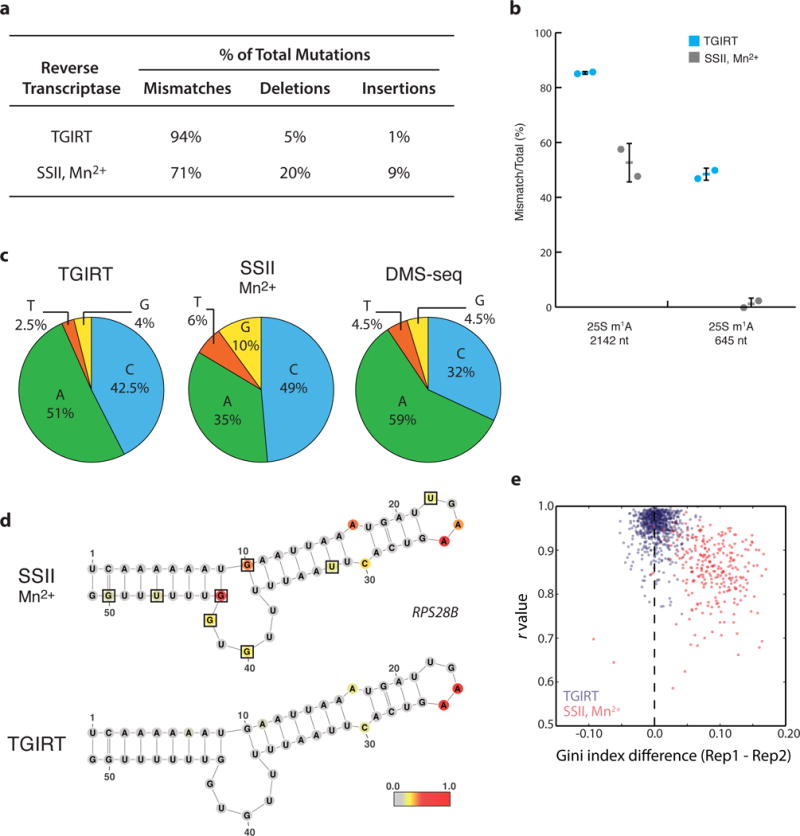
a, Distribution of mutation type generated by SSII/Mn2+ or TGIRT reverse transcription from in vivo DMS-treated yeast mRNA. b, Endogenous m1A modifications in yeast 25S rRNA transcript reveal superior modification detection with TGIRT. Average percent modification (bar) detected at the position across two biological DMS-treated replicates (circles) with error bars representing standard deviation from the average. c, Nucleotide composition of mismatches from TGIRT or SSII/Mn2+ approaches. d, Yeast RPS28B mRNA positive control structure with nucleotides colored by DMS reactivity in vivo. Black boxes outline G/U bases with high background signal. DMS reactivity was calculated as the average ratiometric DMS signal per position across two biological replicates normalized to the highest number of reads in displayed region, which is set to 1.0. e, Genome-wide DMS-MaPseq replicates compared by Pearson’s r value and Gini index for yeast mRNA regions (requiring 15x coverage, resulting in 733 and 272 regions displayed for TGIRT and SSII/Mn2+, respectively).
