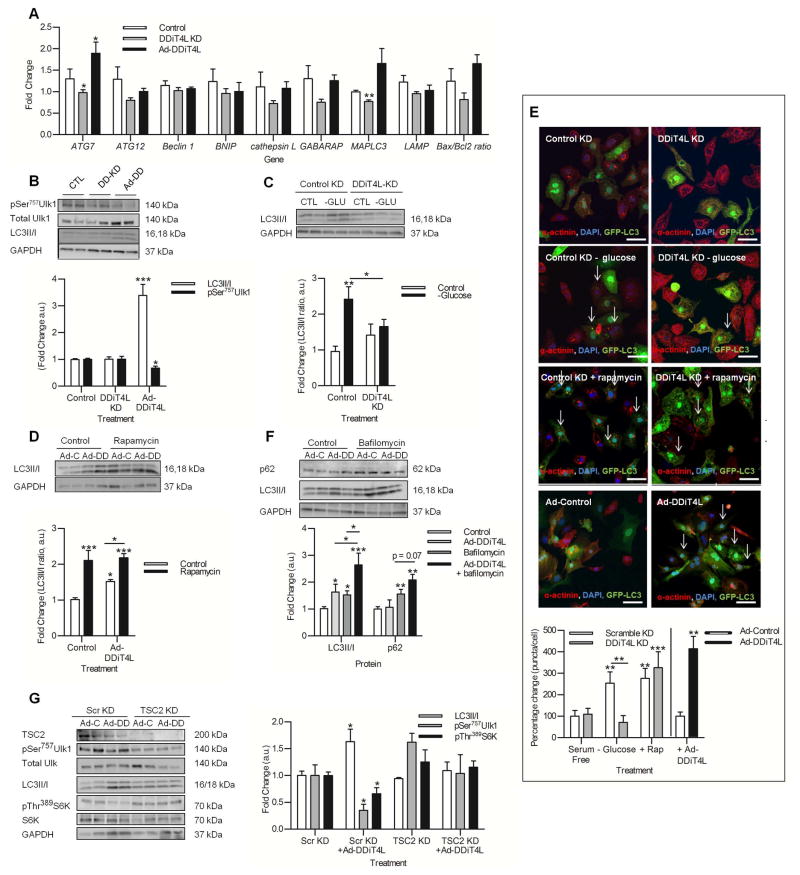
Figure 3. DDiT4L increases autophagic flux in an mTOR-dependent and TSC2-independent manner.
(A) The expression of the autophagy genes ATG7 and MAPLC3 was increased with Ad-DDiT4L infection and decreased with DDiT4L knockdown. n=4 preparations, 2 samples per preparation. Student t-test *p<0.05 compared to control.
(B) Ad-DDiT4L infection (Ad-DD) increased the LC3II/I ratio and decreased phosphorylation of Ser757 in Ulk1 with no changes seen with DDiT4L knockdown (DD-KD). n=4 preparations, 2 samples per preparation. One Way ANOVA followed by Bonferroni posttest *p<0.05, ***p<0.001 compared to control. Phospho-proteins were normalized to total protein.
(C) Glucose deprivation (-GLU) increased autophagy, which was inhibited by DDiT4L knockdown. n=3 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni post-test, *p<0.05, **p<0.01 compared to control or indicated sample.
(D) Treatment of Ad-DDiT4L-infected (Ad-DD) cells with rapamycin did not lead to a further increase in autophagy. n=4 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni post-test *p<0.05, ***p<0.001 compared to control or indicated sample.
(E) Glucose deprivation, rapamycin and Ad-DDiT4L infection increased the number of GFP-LC3 autophagosomes in NRVMs, and knockdown of DDiT4L prevented the increase caused by glucose deprivation. n=4 preparations, 2 samples per preparation, 4 images per coverslip, 100–160 cells per treatment. Arrows indicate cells with autophagosomes. One-Way ANOVA, Bonferroni posttest, *p<0.05, **p<0.01, ***p<0.001 compared to control or indicated sample.
(F) Ad-DDiT4L infection and bafilomycin increased the LC3II/I ratio in an additive fashion. n=6–10 preparations, 2 samples per preparation. Bafilomycin treatment, but not Ad-DDiT4L infection alone, significantly increased p62 abundance. n = 4 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni posttest *p<0.05, **p<0.01, ***p<0.001 compared to control or indicated sample.
(G) TSC2 knockdown attenuated the induction of autophagy by Ad-DDiT4L infection in NRVMs as assessed by LC3II/I or phosphorylation (p) of Ser757 in Ulk1 and inhibited the decrease in pS6K. n=4 preparations, 2 samples per preparation. One-Way ANOVA, Bonferroni post-test *p<0.05 compared to control or indicated sample. Phospho-proteins were normalized to total protein.