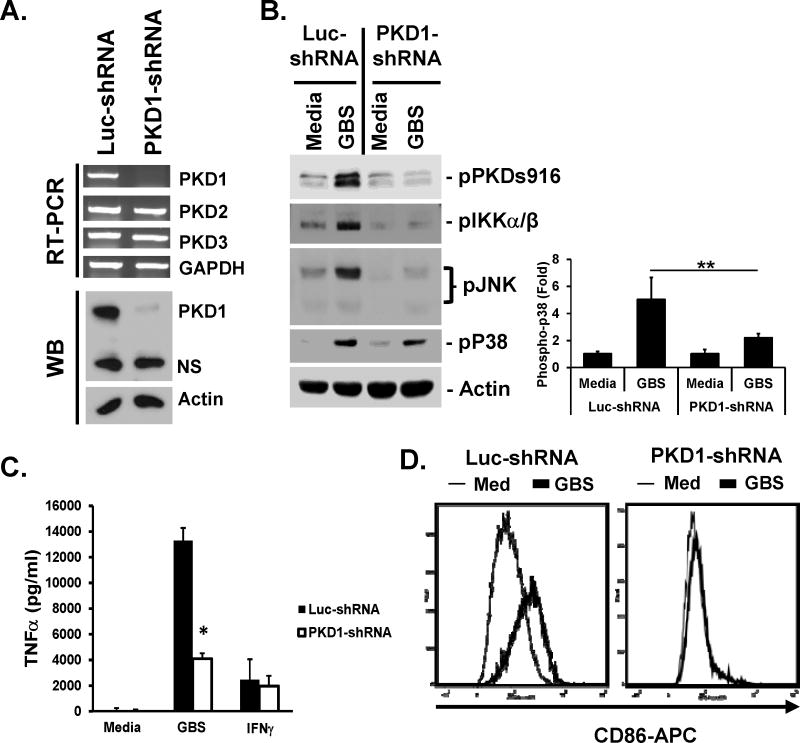
Figure 5. Activation of MAPKs and IKKα/β and production of proinflammatory cytokines induced by GBS is dependent on PKD1 in vitro.
(A) Messenger RNA levels of PKD family members and protein levels of PKD1 in control luciferase-knockdown RAW264.7 cells (Luc-shRNA) and PKD1-knockdown RAW264.7 cells (PKD1-shRNA) were analyzed by RT-PCR and Western blot (WB) assay, respectively. GAPDH and Actin were used as loading controls. (B) (Left panel) Control luciferase-knockdown or PKD1-knockdown RAW264.7 cells were stimulated with medium, live GBS (5 × 107 cfu/ml) or IFNγ (10 ng/ml) for 45 min. Phosphorylation status or expression levels of the indicated proteins were detected by Western blot assay. Actin was used as a loading control. (Right panel) Quantitation of phosphor-p38 in Western blots by densitometry. The density of phosphor-p38 band in each sample was quantitated three times by densitometry and normalized to the density of the actin band in the same sample. Data represent the mean (fold induction from the normalized densitometric value of phosphor-p38 band of the unstimulated Luc-shRNA control sample) ± S.D. from two separate experiments. (C) Control luciferase-knockdown or PKD1-knockdown RAW264.7 cells (1 × 106 cells/ml) were stimulated with medium, GBS (5 × 106 cfu/ml) or IFNγ (10 ng/ml) for 24 hr in the complete DMEM supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin. Levels of TNFα in culture supernatants were analyzed by ELISA. Data represent the mean concentration (pg/ml) ± SD of triplicates. *, p < 0.05. (D) Control luciferase-knockdown or PKD1-knockdown RAW264.7 cells were stimulated with medium, GBS (108 cfu/ml) for 24 hr in the complete DMEM supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin. Surface expression levels of CD86 were detected by flow cytometric analysis. All experiments were repeated two to four times with similar results.