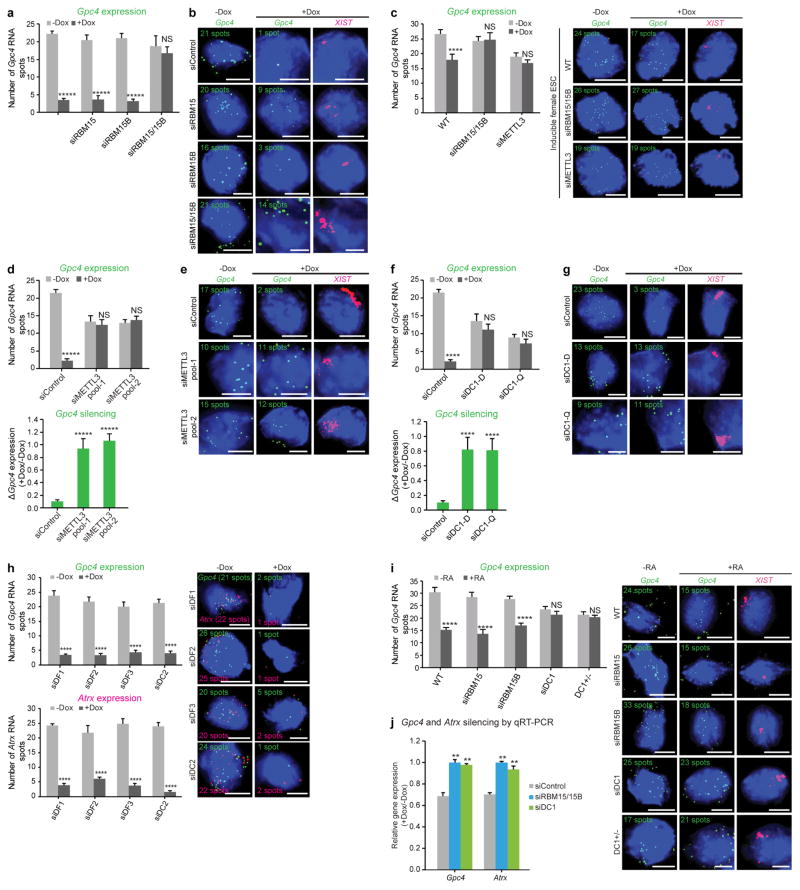
Extended Data Figure 2. Quantification of X-linked gene silencing upon knockdown of m6A readers and writers.
a, b, Quantification of Gpc4 spots upon Rbm15 and Rbm15b knockdown (Fig. 1b, c). The number of Gpc4 spots before and after XIST induction (−Dox and +Dox, respectively) (a). Representative RNA-FISH images with DAPI-stained nuclei with Gpc4 spots (green) and XIST staining (pink, last column) are shown (b). The number of Gpc4 spots is indicated on each FISH image. Scale bar, 5 μm. Data in a are mean ± s.e.m. NS, not significant; *****P < 0.0001 relative to Dox-deficient control by unpaired two-sample t-test. c, m6A modification is necessary for XIST-mediated gene silencing in female pSM33 cells. Quantification of Gpc4 RNA spots with and without induction of XIST expression (left). Representative RNA-FISH images showing Gpc4 RNA spots (green) with DAPI-stained nuclei (right). Wild-type (WT) cells show a normal XIST-induced silencing whereas Gpc4 spots are partially reduced (24 to 17 spots). Similar to male ES pSM33 cells, female ES cells fail to show XIST-mediated gene silencing upon knockdown of Rbm15/15b or Mettl3. Error bars mean ± s.e.m. for 50 cells per sample. NS, not significant; ****P < 0.0001, relative to no-doxycycline control by unpaired two-sample t-test. d, e, Similar to Fig 3c, d, shown is an siRNA pool that targets a (different) region on Mettl3. The data from Fig. 3c, d for the siRNA pool 1 is also shown here for comparison. In both the siControl and siMETTL3-transfected cells, XIST shows aggregation consistent with its interaction with the X chromosome. Thus, early steps of XIST interaction with the X chromosome may not require m6A. Gpc4 counts (d, top) and the change in transcription, as measured by the ratio of Gpc4 +Dox/−Dox. Notably, there is a reduction in Gpc4 and Atrx spots (see Fig. 3d) in siMETTL3-transfected cells, even in the absence of XIST expression. Representative FISH images with DAPI nuclear stain in blue, Gpc4 in green and XIST in pink (e). Following Dox treatment, the number of Gpc4 spots is markedly reduced in the control-transfected cells. However, after knockdown of Mettl3, the number of Gpc4 mRNA spots remain unchanged. Scale bars, 5 μm. Data in c are mean ± s.e.m.across 50 cells. NS, not significant; *****P < 0.0001 relative to no-doxycycline control (top graph) and siControl (bottom graph) by unpaired two-sample t-test. f, g, Similar to d and e, we show a defect in XIST-mediated silencing upon silencing of Ythdc1 as shown in Fig. 4d, e using multiple siRNA pools from different vendors. Targeting a different region of DC1 using a siRNA pool (siDC1-Q) prevents XIST-mediated gene silencing. The data from Fig. 4d, e for the Dharmacon siRNA pool is shown alongside. Data in f are mean ± s.e.m across 50 cells. NS, not significant; ****P < 0.005 relative to no-doxycycline control (top graph) and siControl (bottom graph) by unpaired two-sample t-test. h, DF1, DF2, DF3 and DC2 do not mediate XIST-mediate gene silencing. Quantification of Gpc4 (top left) and Atrx (bottom left) RNA-FISH spots is shown. Representative FISH images with DAPI-stained nuclei (blue) with Gpc4 (green) and Atrx (red) spots are shown (right). The number of detected RNA spots for both the genes are indicated on each FISH image. Scale bars, 5 μm. Data are mean ±s.e.m. across 50 cells from one experiment. ****P < 0.0001 relative to control (−Dox) by unpaired two-sample t-test. i, RBM15/15B and DC1 mediate XIST-mediate gene silencing in differentiating wild-type female ES cells. Quantification of Gpc4 RNA expression was performed in female mouse ES cells in response to retinoic acid-induced (+RA) differentiation by RNA-FISH (left). Representative FISH images showing DAPI-stained nuclei (blue), Gpc4 RNA (green), and XIST (pink) are shown (right). Wild-type cells exhibit normal Gpc4 silencing in response to retinoic acid treatment. Single knockdown of either Rbm15 or Rbm15b also exhibited normal silencing of Gpc4. Double knockdown resulted in no XIST expression (C.-K.C. and M.G., data not shown), reminiscent of the lack of XIST expression in METTL3-deficient ES cells45. CRISPR-mediated homozygous knockout of DC1 (Ythdc1−/ −) cells could not be recovered, suggesting that deletion of this gene is lethal. However, heterozygous knockout of DC1 (Ythdc1−/+) impaired Gpc4 silencing in response to retinoic acid in these cells. These data support the idea that DC1 is required for silencing of X-linked genes during ES cell differentiation. ****P < 0.0001 relative to control by unpaired two-sample t-test. j, qRT–PCR-based validation of effects of RBM15/15B and DC1 on XIST-mediated gene silencing. Gene expression level after XIST induction (+Dox) was normalized to Gapdh before XIST induction (−Dox) in both the siControl and siRbm15/siRbm15b double-knockdown sample. Quantification of the change in gene transcript levels upon expression of XIST is shown for Gpc4 and Atrx. Dox-induced XIST expression led to reduced transcription of both the genes in Control knockdown cells. However, Rbm15 and Rbm15b double knockdown and DC1 knockdown failed to show XIST-induced silencing. **P < 0.01 relative to siControl-transfected cells by unpaired two-sample t-test.