During embryo morphogenesis, minor epithelia are generated after, and then form bridges between, major epithelia (e.g., epidermis and gut). In Caenorhabditis elegans, this delay is regulated by four proteins that control production and localization of polarity proteins: the pioneer factor PHA-4/FoxA, kinesin ZEN-4/MKLP1, its partner CYK-4/MgcRacGAP, and PAR-6.
Abstract
To establish the animal body plan, embryos link the external epidermis to the internal digestive tract. In Caenorhabditis elegans, this linkage is achieved by the arcade cells, which form an epithelial bridge between the foregut and epidermis, but little is known about how development of these three epithelia is coordinated temporally. The arcade cell epithelium is generated after the epidermis and digestive tract epithelia have matured, ensuring that both organs can withstand the mechanical stress of embryo elongation; mistiming of epithelium formation leads to defects in morphogenesis. Using a combination of genetic, bioinformatic, and imaging approaches, we find that temporal regulation of the arcade cell epithelium is mediated by the pioneer transcription factor and master regulator PHA-4/FoxA, followed by the cytoskeletal regulator and kinesin ZEN-4/MKLP1 and the polarity protein PAR-6. We show that PHA-4 directly activates mRNA expression of a broad cohort of epithelial genes, including junctional factor dlg-1. Accumulation of DLG-1 protein is delayed by ZEN-4, acting in concert with its binding partner CYK-4/MgcRacGAP. Our structure–function analysis suggests that nuclear and kinesin functions are dispensable, whereas binding to CYK-4 is essential, for ZEN-4 function in polarity. Finally, PAR-6 is necessary to localize polarity proteins such as DLG-1 within adherens junctions and at the apical surface, thereby generating arcade cell polarity. Our results reveal that the timing of a landmark event during embryonic morphogenesis is mediated by the concerted action of four proteins that delay the formation of an epithelial bridge until the appropriate time. In addition, we find that mammalian FoxA associates with many epithelial genes, suggesting that direct regulation of epithelial identity may be a conserved feature of FoxA factors and a contributor to FoxA function in development and cancer.
INTRODUCTION
Animal embryos coordinate the morphogenesis of their body and internal organs in a process that requires epithelia. Communication between mesenchyme and epithelia is necessary for morphogenesis of the mammalian kidney and lung, for example, as well as the Drosophila trachea (reviewed in Hogan and Kolodziej, 2002; Caviglia and Luschnig, 2014; Combes et al., 2015; McCulley et al., 2015). There is also synergy between different organs that helps shape the embryonic body plan. In Caenorhabditis elegans, the embryo elongates fourfold to generate a long, thin worm from an oblong ball of cells (reviewed in Chisholm and Hardin, 2005; Vuong-Brender et al., 2016). Elongation of the body is coordinated with that of the gut by physically linking the outer epidermis to the inner digestive tract. At the anterior, the link is constructed by the arcade cells, which generate an epithelial bridge between the epidermis and foregut just before body elongation. Proper morphogenesis depends critically on this linkage. Disruption of arcade cell polarization by mutation or laser ablation blocks attachment of the gut tube to the epidermis (Heid et al., 2001; Portereiko and Mango, 2001; Portereiko et al., 2004; Mango, 2009; Kuzmanov et al., 2014). As a consequence, the foregut fails to elongate, and affected animals cannot feed (Portereiko et al., 2004). Similarly, in sma-1 mutants, the foregut attaches to the epidermis, but the embryo body fails to extend fully, and this leads to defects in foregut morphogenesis (McKeown et al., 1998). These phenotypes indicate that proper gut morphogenesis requires attachment to the elongating epidermis. Conversely, if the arcade cells generate an epithelial bridge before epidermal maturation, the digestive tract distorts the morphology of the nose and produces feeding defects (Kelley et al., 2015). These observations illustrate that embryos rely on epithelia to coordinate body morphogenesis and raise the question of how cells dictate the timing of epithelium formation and attachment.
Epithelia become polarized with distinct apical and basolateral domains separated by a junctional domain (Nelson et al., 2013; Roignot et al., 2013). After cell contact–mediated cues, three major polarity complexes are recruited to the membrane of fly and vertebrate epithelia and partition the cell into apical and basolateral domains (reviewed in St Johnston and Ahringer, 2010; Roignot et al., 2013; Rodriguez-Boulan and Macara, 2014). The Crumbs complex and the partitioning-defective (PAR) complex are important for apical identity and/or junction formation (Wodarz et al., 1993, 1995; Gao et al., 2002; Hurd et al., 2003; Roh et al., 2003; Harris and Peifer, 2004; Lemmers et al., 2004; Chen and Macara, 2005; St Johnston and Ahringer, 2010; Roignot et al., 2013; Rodriguez-Boulan and Macara, 2014). The basolateral surface is marked by the Scribble complex, which contains Scribble, Discs Large (DLG), and Lethal Giant Larvae (St Johnston and Ahringer, 2010; Roignot et al., 2013). Diffusion between the apical and basolateral domains is blocked by the junctional domain, which also provides tissue integrity via cell–cell adhesion (Hartsock and Nelson, 2008). The combined actions of these polarity modules establish and maintain polarity in a range of tissues.
In the nematode, all epithelial cells form via mesenchymal-to-epithelial transition (MET) from unpolarized precursors (Portereiko et al., 2004; Achilleos et al., 2010). C. elegans epithelia have a single electron-dense junction, termed the C. elegans apical junction (CeAJ; McMahon et al., 2001), which has properties of both tight and adherens junctions (Pásti and Labouesse, 2014). The apicobasal distribution of factors, including those in the Crumbs, Par, and Scribble groups, is similar between worm and vertebrate epithelia (Pásti and Labouesse, 2014). The three Crumbs homologues localize to the apical surface (Bossinger et al., 2001; Segbert et al., 2004; Waaijers et al., 2015), PAR-6 localizes to the apical domain (Leung et al., 1999; McMahon et al., 2001; Nance et al., 2003), and DLG-1 is found in the basal half of the CeAJ (Bossinger et al., 2001; Firestein and Rongo, 2001; Koppen et al., 2001; McMahon et al., 2001; Segbert et al., 2004). Distinct epithelia rely on the same cohort of epithelially expressed genes, indicating that temporal control cannot be explained by distinct, paralogous factors acting in different organs. For example, par-6 and dlg-1 are critical for the integrity of adherens junctions in all tested organs, and mutants for either gene fail to elongate their body or digestive tract (Bossinger et al., 2001; Firestein and Rongo, 2001; Koppen et al., 2001; McMahon et al., 2001; Totong et al., 2007).
Transcription factors can drive both MET and the reverse process, epithelial-to-mesenchymal transition (EMT; Chaffer et al., 2007; Lamouille et al., 2014). Targets of EMT include E-cadherin and integrin (Batlle et al., 2000; Cano et al., 2000; Comijn et al., 2001; Perez-Moreno et al., 2001; Hajra et al., 2002; Bolós et al., 2003; Yang et al., 2004, 2009; Eger et al., 2005), but the direct transcription factor targets that mediate MET are largely unknown (Chaffer et al., 2007). In this study, we focus on forkhead box transcription factor subgroup A (FoxA) factors, which promote the development of several epithelial organs and are often found in tumors derived from epithelia (Friedman and Kaestner, 2006). For example, mammalian FoxA1 and FoxA2 are expressed in the lung, thyroid, kidney, and pancreas, and FoxA1 is also found in the mammary gland (Besnard et al., 2004; Friedman and Kaestner, 2006). C. elegans has one FoxA factor, pha-4, which functions in two epithelial organs, the gonad and foregut (Mango et al., 1994; Updike and Mango, 2007). In mammals, reduction of FoxA can promote EMT in lung, liver, prostate, and breast by reducing expression of Slug, E-cadherin, or MMP-9 (Tang et al., 2011; Wang et al., 2014; Zhang et al., 2015). However, homologues of Slug, E-cadherin, and metalloproteases play no known role in polarizing epithelia in worms (Costa et al., 1998; Raich et al., 1999; Fraser et al., 2000; Kamath et al., 2003; Simmer et al., 2003; Rual et al., 2004; Sönnichsen et al., 2005; Von Stetina and Mango, 2015), indicating that either FoxA factors have additional, conserved target genes or regulate distinct epithelial genes in different organs and/or species. Here we define direct targets of C. elegans FoxA and explore their orthologues in vertebrates.
In this work, we examine C. elegans epithelium formation during morphogenesis, when a ball of cells is transformed into a long, thin worm. We find that epithelia are generated just before the onset of their associated morphogenetic event. We focus on the arcade cells, which form an epithelium that bridges the epidermis and foregut during late embryogenesis. A core set of epithelial factors is activated by the pioneer factor defective pharynx development 4 (PHA-4)/FoxA, but protein accumulation and localization are delayed by zygotic enclosure defective 4 (ZEN-4)/MKLP1, cytokinesis defective 4 (CYK-4)/MgcRacGAP, and PAR-6. We extend these results to FoxA factors in mammalian tissues and determine that vertebrate FoxA factors bind many orthologous target genes. The results reveal how the exquisite timing of embryonic morphogenesis depends on temporally coordinated regulation of a common core of epithelial factors at the RNA and protein levels.
RESULTS
Overview of C. elegans epithelium formation
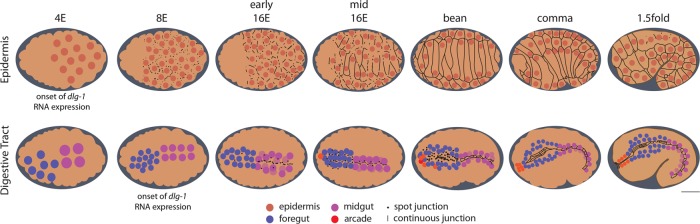
Timing of embryo development can be tracked by the number of E (endodermal) cells and by embryo shape (Figure 1; Sulston et al., 1983; Leung et al., 1999). The epidermis is the first epithelium to polarize, during the late 8E stage (∼200 cells; Sulston et al., 1983; Podbilewicz and White, 1994; Leung et al., 1999; McMahon et al., 2001; Chisholm and Hardin, 2005). This period occurs soon after the epidermal precursors are born between the 2E and mid-8E stages and before epiboly at the bean stage (Sulston et al., 1983). Once the epithelium has formed, it migrates over the lateral and ventral surfaces of the embryo, enclosing the embryo in preparation for body elongation (Priess and Hirsh, 1986; Williams-Masson et al., 1997; Figure 1).
FIGURE 1:
C. elegans embryonic stages and epithelial cell anatomy. Anterior is left. Top, epidermis; bottom, digestive tract. Nuclei of the epidermis (orange), foregut (blue), midgut (magenta), and arcade cells (red). Staging is determined by the number of midgut (or E) cells for early embryos and embryo shape at late stages. Junctional proteins (e.g., DLG-1/Discs large, black) become apparent in the epidermis in the 8E stage as spot junctions, which become larger in the early 16E and resolve into continuous junctions by the mid-16E stage. By the 1.5-fold stage, some epidermal cells fuse, creating large, multinucleate cells. The digestive track polarizes in a posterior-to-anterior direction, with the midgut expressing junctional protein at the early 16E stage, followed soon thereafter by the foregut at the mid 16E stage. Again, spot junctions precede continuous junctions. The midgut transitions by the bean stage, and the foregut by the comma stage. The nine arcade cells are born at the mid 16E stage (only six are drawn). These cells cluster together anterior to the foregut by the comma stage but do not express junctional protein until they polarize between the comma and 1.5-fold stages. The onset of dlg-1 RNA expression is indicated for the epidermis (4E) and foregut/midgut (8E). The arcade cells express dlg-1 RNA from their birth in the 16E stage. Scale bar, 10 μm. Embryo length to scale, but nuclear size is not necessarily to scale.
The digestive tract polarizes progressively, with midgut epithelialization commencing at the 8E stage and junction formation starting in the early 16E stage, whereas the foregut shows the first hallmarks of polarity at early 16E and begins to form junctions in mid-16E (Figure 1; Totong et al., 2007; Achilleos et al., 2010; Rasmussen et al., 2012). Between the comma and 1.75-fold stages, the anteriorly located arcade cells and posterior hindgut undergo MET to generate two small epithelia that link the epidermis to the foregut and hindgut, respectively. The result is a torus with the epidermis on the outside and the digestive tract on the inside, held under tension by its distal attachments. The torus elongates fourfold to generate a worm with an extended, linear digestive tract (Chisholm and Hardin, 2005; Vuong-Brender et al., 2016). Without mechanical coupling of the gut to the epidermis, the digestive tract cannot produce its proper elongated form (Portereiko and Mango, 2001; Mango, 2009; Kelley et al., 2015).
Asynchronous accumulation of dlg-1 RNA and protein in different organs
To understand the temporal regulation of epithelium formation, we determined the onset of expression for polarity factors by surveying members of the Par (par-6) and Scribble (dlg-1) modules with single-molecule fluorescence in situ hybridization (smFISH) for RNA (Ji and van Oudenaarden, 2012). PAR-6 plays an early and key role in maturing apical junctions within the epidermis and digestive tract (Totong et al., 2007), whereas DLG-1 is one of the last components added to epithelial junctions and is essential for their maturation (Totong et al., 2007). We found that par-6 RNA was contributed maternally, as predicted from prior studies (Watts et al., 1996; Nance et al., 2003), and no increase due to zygotic par-6 RNA was detected (Supplemental Figure S1; Totong et al., 2007). dlg-1 was induced zygotically, with RNA accumulating in different organs at different times, before the generation of each epithelium (described later). We also assayed the onset of protein expression, as this demonstrates when the epithelium is in the final stages of maturation. Whereas the onset of DLG-1 protein has been documented for the epidermis (Podbilewicz and White 1994; Bossinger et al., 2001; McMahon et al., 2001; Chisholm and Hardin 2005) and midgut (Leung et al 1999; Totong et al., 2007; Achilleos et al., 2010), it has not been described for the foregut, and is unclear how the onset of expression compares among these three epithelia.
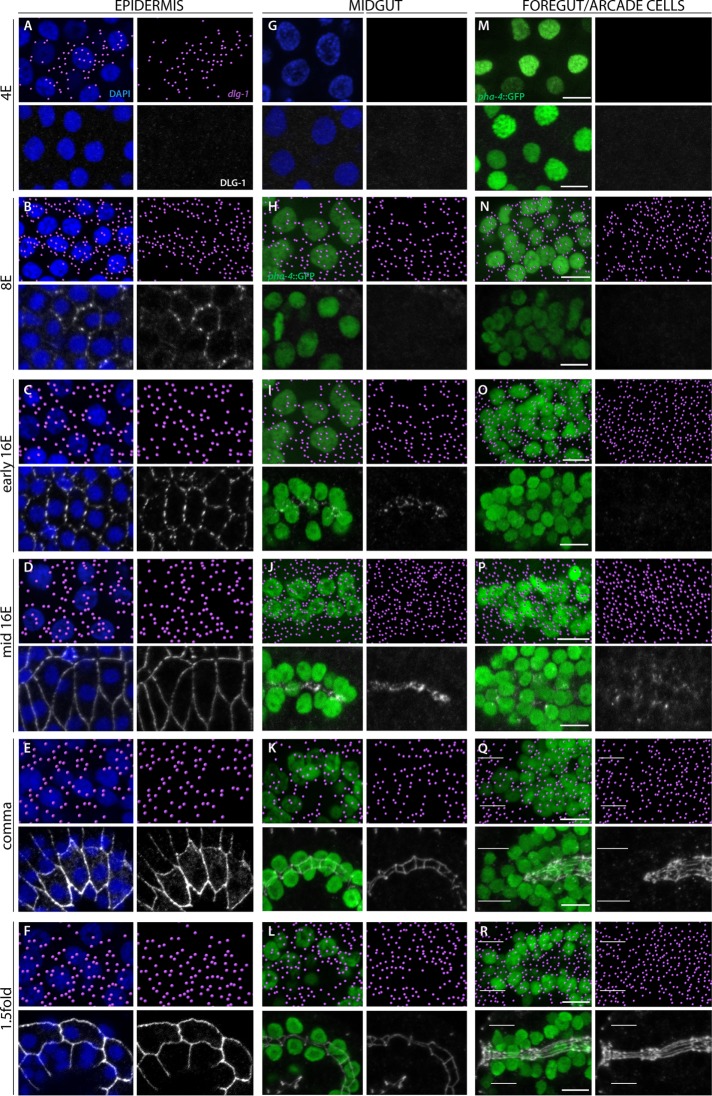
The epidermis was the first epithelium to express dlg-1 mRNA. It was initially detected at the late 4E stage but with no detectable DLG-1 protein (Figures 1 and 2A). The level of dlg-1 mRNA increased during the 8E stage (Figure 2B) and was maintained throughout the 16E and elongation stages (comma, 1.5-fold; Figure 2, C–F). DLG-1 protein was first observed during the late 8E stage, with puncta of protein visible on the membrane of nascent epidermal cells (Figure 2B). These puncta began to coalesce at the early 16E stage (Figure 2C) and formed a continuous, circumferential junction by the mid-16E stage (Figure 2D). The level of DLG-1 increased during the elongation stages (comma, 1.5-fold; Figure 2, E and F), as the cells changed shape to convert the embryo from a ball into a vermiform.
FIGURE 2:
Onset of dlg-1 RNA and protein expression in epithelia. dlg-1 RNA is pseudocolored magenta (top); DLG-1 protein is labeled in white (bottom). RNA and protein data are from different embryos. Nuclei of the epidermis (A–F) and 4E midgut (G) are visualized by DAPI (blue). PHA-4::GFP (green) marks the midgut (H–L), foregut (M–R), and arcade cells (white bars in Q and R). (A–F) dlg-1 RNA is first detected in the 4E epidermis (A), whereas the onset of protein expression is at the 8E stage (B). RNA and protein are continuously expressed throughout the 16E and morphogenetic stages (C–F). Note that the protein first forms spot junctions (B, C), which resolve into continuous bands (D–F). (G–L) The midgut first expresses RNA at the 8E stage and protein at the early 16E. The protein first forms spot junctions (I), which quickly resolve into continuous bands (J). The RNA and protein are also detected during morphogenesis (K, L). (M–R) Like the midgut, RNA expression in the foregut begins at the 8E stage (N). Unlike the midgut, protein expression is first visible as spots by the mid rather than the early 16E stage (compare I vs. O and P). Foregut cells have formed continuous junctions by the comma stage (Q). The arcade cells (between the white bars) have RNA but no protein at the comma stage (Q), and between the comma and 1.5-fold, the arcade cells express protein and form continuous junctions (R). All images are maximum intensity projections. Anterior is to the left. Scale bars, 5 μm. Embryos were fixed and either hybridized with probes to detect RNA expression or stained with antibodies to detect protein. The midgut and foregut/arcade cell images are from the same embryo; the epidermis images at the same stage are from different embryos. smFISH data were imported into Imaris 3D software (Materials and Methods).
The digestive tract began to express dlg-1 mRNA at the 8E stage (Figure 2, H and N), and, similar to the epidermis, the levels of RNA increased throughout the 8E and 16E stages (Figure 2, I, J, and O–P). DLG-1 protein was first observed in midgut precursors at the early 16E stage (Figure 2I), where puncta of protein appeared at the lateral surface and rapidly coalesced at the apical surface, in agreement with previous studies (Supplemental Figure S2; Leung et al., 1999; Totong et al., 2007; Achilleos et al., 2010). By the mid-16E stage (∼20–40 min later), the puncta of DLG-1 had banded together to form cell junctions (Figure 2J), which continued to expand and mature as the embryo elongated (comma, 1.5-fold stages; Figures 1 and 2, K and L). The RNA remained expressed in the intestine throughout all of these stages (Figure 2, I–L).
In the foregut, DLG-1 protein was first detectable by the mid-16E stage (Figure 2P), suggesting that translation of dlg-1 mRNA was delayed in this tissue by ∼20–40 min. We observed membrane-associated DLG-1 puncta on cell surfaces throughout the foregut at the 16E stage (Supplemental Figure S2). These spots accumulated at the nascent apical surface by the bean stage (Supplemental Figure S2), where they joined together to form connected junctions by the comma stage (Figures 1 and 2Q). The RNA remained expressed throughout these stages (Figure 2, O–R). Together, these data show that the digestive tract forms in a piecemeal manner, with the midgut polarizing and maturing before the foregut, and both after the epidermis.
By the end of the comma stage, the epithelial cells of the foregut and midgut were linked together via adherens junctions, yet the digestive tract remained isolated from the epidermis. The digestive tract became attached to the epidermis at the anterior via epithelialization of the arcade cells (Figure 1; Portereiko and Mango, 2001). The arcade cells are born during the mid-16E stage, starting ∼290 min after the first division (Sulston et al., 1983). The majority of these cells are anterior to the foregut primordium and express dlg-1 mRNA from birth (Figure 2O). DLG-1 protein accumulated ∼100 min later in the arcades (Figure 2R), after the epidermis and foregut had both formed epithelia and soon before the arcade cells became an epithelium (i.e., between the comma and 1.25-fold stage, ∼390–400 min after the first division; Portereiko and Mango, 2001; Portereiko et al., 2004). The presence of RNA but lack of protein was detectable by the 16E stage (Supplemental Figure S2C) but was clearest at the comma stage, when the arcade cells clustered together as a group anterior to the foregut epithelium (Figure 2Q). Thus there was a delay in protein accumulation, suggesting that either the RNA was translationally repressed or protein was made but degraded immediately. These results reveal that dlg-1 RNA and protein are uncoupled in different organs, with the protein appearing just before polarity establishment. Figure 1 summarizes the onset of RNA and protein expression presented here.
PHA-4 activates dlg-1 in the foregut and arcade cells
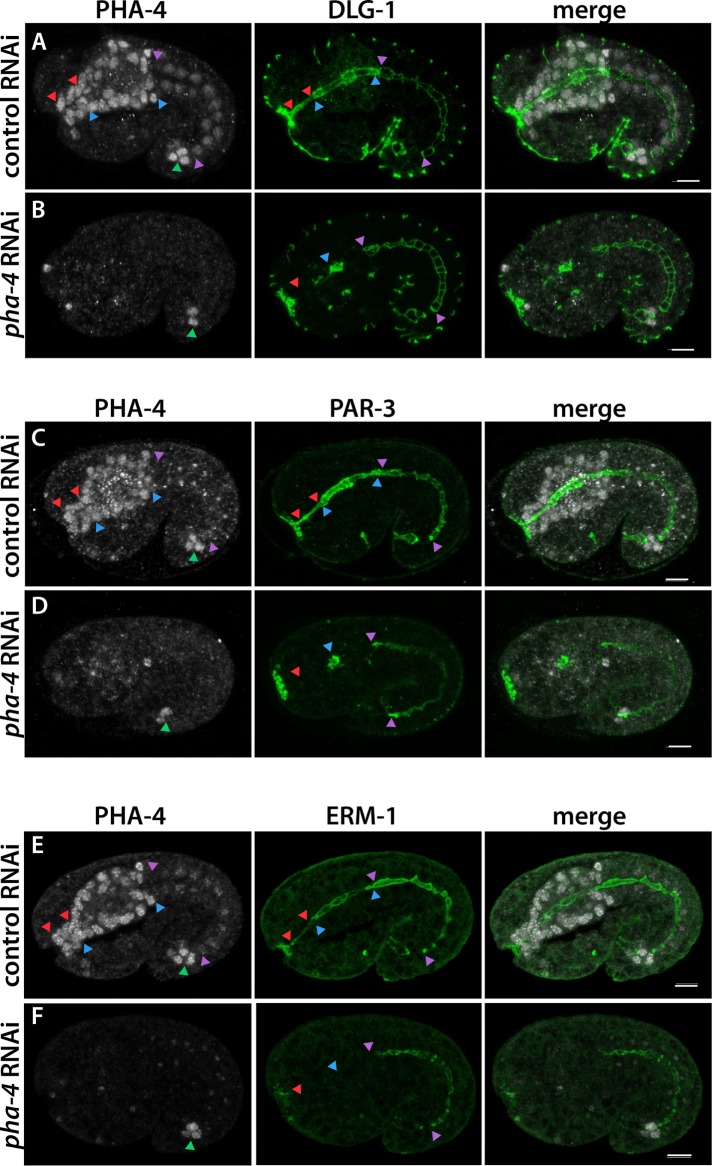
The onset of dlg-1 RNA expression suggested a transcriptional component to temporal regulation of epithelium formation. One central regulator of the digestive tract is PHA-4, a pioneer transcription factor that is also the master regulator for the C. elegans foregut (Horner et al., 1998; Gaudet and Mango, 2002; Mango, 2009; Hsu et al., 2015). To determine whether PHA-4 was important for dlg-1 expression, we performed RNA interference (RNAi) against pha-4 and observed reduced DLG-1 in the foregut and arcade cells but normal expression in the midgut and epidermis (Figure 3A). Antibody staining against endogenous PHA-4 confirmed that RNAi was effective (Figure 3B). Apical factors PAR-3 and ezrin/radixin/moesin homologue 1 (ERM-1) were similarly disrupted by loss of pha-4 (Figure 3, C–F), suggesting widespread disruption of epithelial factors.
FIGURE 3:
PHA-4/FoxA is required for polarity protein expression in the foregut. The arcade cell epithelium lies between red arrowheads, the foregut epithelium between blue arrowheads, and the midgut epithelium between purple arrowheads, and hindgut cells are marked by a green arrowhead. (A, C, E) In control RNAi embryos, PHA-4 (white) is strongly expressed in arcade, foregut, and hindgut cells and dimly expressed in the midgut. Markers of polarity (green), such as DLG-1 (A), PAR-3 (C), and ERM-1 (E), are detected at the apical or junctional domains of the digestive tract. (B, D, F) After pha-4 RNAi, PHA-4 is undetectable in the arcade, foregut, or midgut cells. The hindgut is resistant to pha-4(RNAi) and serves as an internal control for antibody staining. In pha-4(RNAi) embryos, polarity protein expression is ablated in arcade cells (red arrowhead) and decreased (DLG-1, PAR-3) or absent (ERM-1) in the foregut. If protein remains, it is detected only in the foregut remnant (blue arrowhead). Expression in the midgut is unchanged (purple arrowheads). All images are maximum intensity projections through the digestive tract. Anterior is to the left. Scale bars, 5 μm.
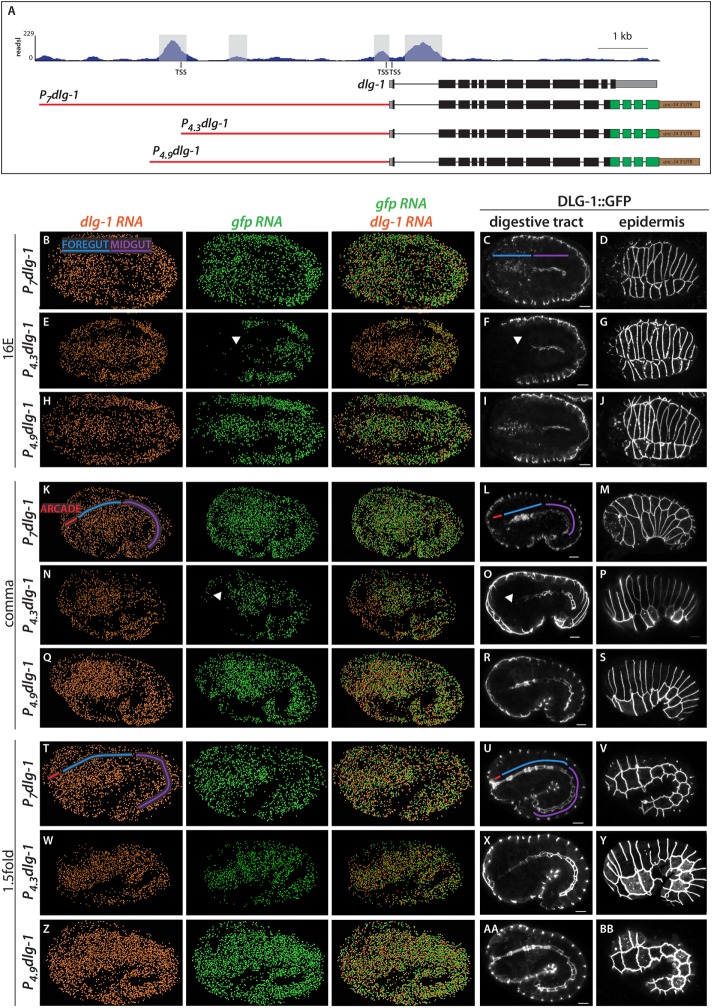
Next we determined whether regulation of dlg-1 transcription by PHA-4 was direct. PHA-4 binding has been mapped by chromatin immunoprecipitation sequencing (ChIP-seq; Zhong et al., 2010). Examination of ChIP-seq data demonstrated two binding sites for PHA-4, one near the upstream dlg-1 transcription start site (TSS; Figure 4A; Saito et al., 2013) and a second site just before the ATG start and within the first intron (Figure 4A). To test whether these sites contributed to dlg-1 expression, we used a DLG-1::green fluorescent protein (GFP) reporter, P7dlg-1, with 7 kb of upstream sequences, which was able to rescue loss of endogenous dlg-1 (McMahon et al., 2001). The reporter expressed DLG-1 appropriately: endogenous dlg-1 and transgenic gfp RNA were both detectable in the foregut and midgut at the 8E stage (i.e., at the onset of endogenous gene expression; Figure 2H and Supplemental Figure S3) and abundant in these tissues by the 16E stage (Figure 4B). Live imaging demonstrated that DLG-1::GFP protein was enriched at the midline of the midgut and clustered into junctions at the 16E stage (Figure 4C). In the mid-16E, DLG-1::GFP was detectable as spots spread throughout the foregut, similar to the endogenous protein (compare Figures 4C and 2P). At the comma stage, both endogenous and transgenic RNAs were detectable throughout the digestive tract (including the arcade cells; Figure 4K), whereas DLG-1::GFP protein, like endogenous DLG-1, was visible only in the foregut and midgut (Figure 4L). By the 1.5-fold stage, DLG-1::GFP was also visible in the arcade cells, as expected (Figure 4U). Thus the P7dlg-1 transgene contained all the information necessary for appropriate temporal expression of RNA and protein in the digestive tract and epidermis (Figure 4, D, M, and V).
FIGURE 4:
Upstream PHA-4 binding site is necessary for the onset of dlg-1 expression in the foregut and arcade cells. (A) PHA-4::GFP ChIP-seq profile at the endogenous dlg-1 locus (top). Only the long dlg-1 isoform is shown. Exons are black boxes, 5′ and 3′ UTR sequence are gray, and introns are black lines. There are four PHA-4 ChIP peaks (highlighted in gray): two surrounding the first exon, one ∼3 kb upstream, and one ∼4.5 kb upstream near the upstream TSS. Three GFP-tagged constructs are also shown, with GFP in green and a heterologous unc-54 3′UTR in brown. Cis-regulatory sequences are red. P7dlg-1 contains 7 kb of sequence, P4.3dlg-1 contains 4.3 kb and includes the upstream TSS but excludes the most upstream PHA-4 peak, and P4.9dlg-1 contains 4.9 kb and includes the most upstream PHA-4 peak. (B–Z) Maximum intensity projections of worm embryos expressing the dlg-1 promoter constructs at the 16E, comma, or 1.5-fold stages. Transgenic embryos were cohybridized with probes to detect dlg-1 (orange) and gfp (green) transcripts via smFISH (columns 1–3). smFISH data were imported into Imaris 3D software (Materials and Methods). DLG-1::GFP protein (white) was detected by live imaging (columns 4 and 5). For every row, DLG-1::GFP images are two focal planes from the same embryo. Note that the RNA and protein data are from different embryos. Areas corresponding to the arcade cells are labeled with a red line, foregut in blue, and midgut in purple. (B, K, T) Embryos expressing DLG-1::GFP under the P7dlg-1 promoter have transgenic gfp mRNA expression that matches endogenous dlg-1 mRNA (compare orange and green mRNA profiles). DLG-1::GFP protein (C, D, L, M, U, V) is similar to endogenous (compare to Figure 2). (E, N, W) Embryos expressing DLG-1::GFP under the P4.3dlg-1 promoter have transgenic gfp mRNA that matches endogenous dlg-1 mRNA in the epidermis and midgut. Expression in the foregut is delayed (white arrowhead). In the 16E stage (E), gfp mRNA is nearly absent in the foregut. By the comma stage (N), most of the foregut has gfp mRNA, but the arcade cells have no expression. Transgenic mRNA expression in the arcade cells and anterior foregut is seen by the 1.5-fold stage (W). DLG-1::GFP protein (F, O, X; white arrowheads point to absent expression in foregut/arcades). (H, Q, Z, AA, BB) Embryos expressing DLG-1::GFP under the P4.9dlg-1 promoter have transgenic mRNA and protein expression profiles that match their endogenous counterparts in all tissues. All images are maximum intensity projections. Anterior is to the left. Scale bars, 5 μm.
We performed promoter deletion analysis to determine whether PHA-4–bound sites contributed to dlg-1 activation. We generated worms expressing two different DLG-1::GFP constructs: one that contained all PHA-4–binding sites (P4.9dlg-1 for 4.9 kb) and one that carried the site near the ATG start but lacked the major binding site at the upstream TSS (P4.3dlg-1 for 4.3 kb; Figure 4A). P4.3dlg-1 faithfully recapitulated the onset of expression in the epidermis (Figure 4, G, P, and Y; unpublished data) and midgut (Figure 4, E, F, N, O, W, and X, and Supplemental Figure S3), similar to endogenous dlg-1. However, P4.3dlg-1 was silent at the 8E (Supplemental Figure S3) and 16E stages in the foregut and arcades, when endogenous dlg-1 was abundant (Figure 4E; compare gfp RNA [green] to dlg-1 RNA [orange]); DLG-1::GFP protein was undetectable (Figure 4F). Expression in the foregut of P4.3dlg-1 transgenic embryos commenced slowly, progressing from posterior to anterior. Specifically, by the comma stage, the foregut (but not the arcade cells) had abundant gfp RNA (Figure 4N), whereas protein was most abundant in the posterior foregut (Figure 4O). At the 1.5-fold stage, transgenic mRNA and protein were detected throughout the digestive tract, although GFP expression was very dim in the arcade cells in most embryos (Figure 4, W and X). GFP was expressed at normal levels in the arcade cells by the twofold stage (unpublished data). Adding back 600 base pairs of sequence (P4.9dlg-1), including the upstream PHA-4–binding site, restored wild-type expression in the foregut (Figure 4, H, I, Q, R, Z, and AA, and Supplemental Figure S3). These data suggest that the PHA-4–binding site near the upstream TSS is critical for onset of expression in the foregut, whereas other PHA-4–binding sites and sites for other factors mediate late foregut expression (or maintenance of expression) and expression in the midgut and epidermis.
In summary, three lines of evidence suggest that PHA-4 contributes to the proper onset of dlg-1 expression: first, DLG-1 expression was reduced or absent from pha-4 mutant foreguts and arcade cells; second, PHA-4 ChIP peaks were detected within dlg-1 regulatory elements (Zhong et al., 2010); and third, deletion analysis implicated the upstream PHA-4–binding site (∼4 kb upstream of the ATG start) for normal onset of expression in the foregut and arcade cells (but not the midgut or epidermis). Thus distinct regulatory elements control the onset of dlg-1 expression in foregut and arcade cells versus midgut and epidermis.
PHA-4 activates an epithelial program during foregut polarization
To extend our observations beyond dlg-1, we examined PHA-4 association with genes encoding other polarity proteins, relying again on the modENCODE PHA-4 ChIP-seq data set (Zhong et al., 2010). Strikingly, genes for every epithelial polarity protein annotated as expressed in the embryonic foregut (27 of 27) had an associated PHA-4 peak, many close to the TSS or within the first large intron, which are common sites for bona fide PHA-4 targets (Gaudet and Mango, 2002; Gaudet et al., 2004; Figure 5 and Table 1). For example, par-3 had two PHA-4 ChIP peaks within the large, first intron (which is a promoter for the smaller, epithelial-specific isoform; Achilleos et al., 2010), erm-1 had an upstream peak and one in a large intron, and the α-catenin hmp-1 contained one peak at the TSS. In support of PHA-4 regulating these epithelial-specific genes, we found reduced levels of ERM-1 and PAR-3 in pha-4(RNAi) mutant foreguts (Figure 3, C–F; antibodies for HMP-1 were not available for testing).
FIGURE 5:
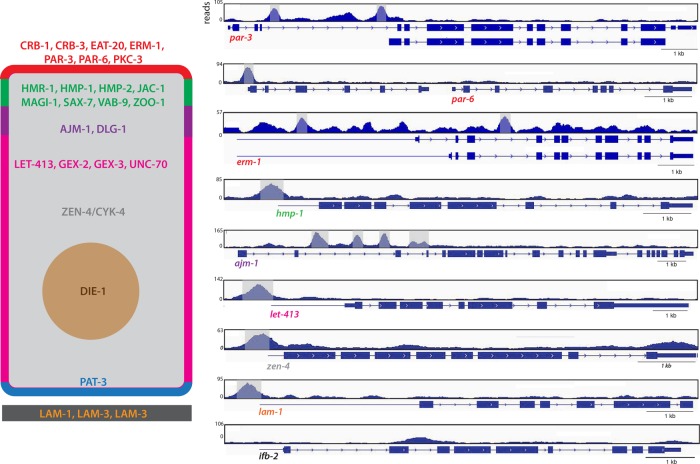
PHA-4 binds to regulatory elements of epithelial factor genes. Left, generic C. elegans epithelial cell, with the apical domain in red, apical junction in green, basal junction in purple, lateral domain in fuschia, basal domain in blue, extracellular matrix in black, cytoplasm in gray, and nucleus in brown. Selected epithelial factors whose regulatory regions are bound by PHA-4 are shown. Right, genome browser displays of PHA-4–binding peaks for a subset of selected genes. Exons are denoted by blue boxes and introns by blue lines with arrowheads. Blue lines preceding the first exon represent untranslated regions. Significant PHA-4 peaks (MACS2; see Materials and Methods) are highlighted in gray. Note that the smaller isoform of par-3 is epithelial specific, suggesting that the large upstream intron in which PHA-4 binds could specifically regulate the expression of this isoform. par-6 is the second gene in an operon, and therefore the binding associated with the upstream gene is also associated with par-6. As a negative control, the midgut specific gene ifb-2 is shown, which lacks PHA-4 binding (bottom).
TABLE 1:
Analysis of foregut epithelial genes for PHA-4 binding.
| Gene | PHA-4 bound? | Location of peak(s) | Expression pattern |
|---|---|---|---|
| afd-1 | Yes | Upstream, at TSS, and/or intronic, depending on isoform | Epithelial |
| ajm-1 | Yes | Intronic | Epithelial |
| cdh-4 | Yes | Intronic | Epithelial |
| crb-1 | Yes | Intronic | Epithelial |
| crb-3 | Yes | Upstream | Epithelial |
| dlg-1 | Yes | Upstream at TSS, intronic | Epithelial |
| eat-20 | Yes | Upstream, at TSS, intronic | Epithelial |
| erm-1 | Yes | Upstream, intronic | Epithelial |
| hmp-1 | Yes | Upstream at TSS | Epithelial |
| hmp-2 | Yes (operon) | Upstream at TSS of operon | Epithelial |
| hmr-1 | Yes | Upstream, at TSS, intronic | Epithelial |
| jac-1 | Yes | Upstream, intronic | Epithelial |
| lam-1 | Yes | Upstream at TSS | Epithelial |
| lam-2 | Yes | Upstream at TSS | Epithelial |
| lam-3 | Yes | Upstream | Epithelial |
| let-413 | Yes | Upstream at TSS | Epithelial |
| let-502 | Yes | Upstream at TSS | Epithelial |
| let-805 | Yes | Upstream and within gene body | Epithelial |
| lgl-1 | Yes | Upstream at TSS | Epithelial |
| magi-1 | Yes | Upstream near TSS, intronic | Epithelial |
| mel-11 | Yes | Upstream TSS | Epithelial |
| par-3 | Yes | Upstream and at TSS of smaller, epithelial isoform | Epithelial |
| par-6 | Yes (operon) | Upstream at TSS of operon | Epithelial |
| pat-3 | Yes | Upstream at TSS | Epithelial |
| pkc-3 | Yes | Upstream at TSS | Epithelial |
| sax-7 | Yes | Upstream, intronic | Epithelial |
| vab-9 | Yes | Upstream, at TSS | Epithelial |
| cdc-42 | Yes | Upstream at TSS | Broad |
| die-1 | Yes | Upstream, at TSS, intronic | Broad |
| ifa-1 | Yes | Upstream at TSS and intronic | Broad |
| ifb-1 | Yes | Intronic | Broad |
| let-858 | Yes | Upstream at TSS | Broad |
| mig-10 | Yes | Upstream/intronic, depending on isoform | Broad |
| pix-1 | Yes | Upstream at TSS, intronic | Broad |
| srgp-1 | Yes | Upstream at TSS | Broad |
| unc-34 | Yes | Within first intron; also one several kilobases upstream | Broad |
| unc-94 | Yes | Within large introns 1 and 3 (or upstream of smaller isoform at TSS) | Broad |
| vab-10 | Yes | Upstream, at TSS, intronic | Broad |
| vab-19 | Yes | Upstream | Broad |
| wsp-1 | Yes | Upstream, intronic | Broad |
| eps-8 | Yes | Upstream, at TSS, intronic | Broad |
| cyk-4 | Yes | Upstream at TSS | Ubiquitous |
| gex-2 | Yes | Upstream at TSS (potentially at bidirectional promoter) | Ubiquitous |
| gex-3 | Yes | Upstream at TSS | Ubiquitous |
| rho-1 | Yes | Upstream at TSS | Ubiquitous |
| spc-1 | Yes | Upstream at TSS | Ubiquitous |
| toca-1 | Yes | Upstream, at TSS, intronic | Ubiquitous |
| unc-70 | Yes | Upstream at TSS, intronic | Ubiquitous |
| wve-1 | Yes | Upstream at TSS (potentially at bidirectional promoter) | Ubiquitous |
| zen-4 | Yes | Upstream at TSS | Ubiquitous |
| abi-1 | Yes | Upstream at TSS (potentially at bidirectional promoter) | Unknown |
| apr-1 | No | n/a | Ubiquitous |
| ced-10 | No | n/a | Ubiquitous |
| ect-2 | No | n/a | Ubiquitous |
| mig-2 | No | n/a | Ubiquitous |
| toca-2 | No | n/a | Ubiquitous |
| clc-1 | No | n/a | Unknown |
| rac-2 | No | n/a | Unknown |
Fifty-seven C. elegans genes that have roles and/or are expressed in epithelial cells, including those of the foregut. Of these, 50 have an associated PHA-4 ChIP peak. The location of the ChIP peaks (the majority of which are at the TSS of the bound genes) and expression pattern of the genes are also shown.
We examined other genes that have important functions in epithelial morphogenesis but a broad expression pattern within the embryo. We found that 20 of 30 genes had PHA-4 peaks associated with cis-regulatory sequences, three of 30 had PHA-4 associated with the TSS of the polarity gene and the TSS of a neighboring gene on the opposite strand in a potentially bidirectional promoter, and seven of 30 had no associated PHA-4 peak (Table 1). On the other hand, we rarely observed PHA-4 associated with genes that are expressed in other epithelia but not in foregut or arcade cells (only one of 12). For example, ifb-2, which codes for an apically localized intermediate filament protein expressed in the midgut (Bossinger et al., 2004), lacked PHA-4 bound to its regulatory sequences (Figure 5 and Supplemental Table S1). These findings suggest that PHA-4 regulates a broad cohort of epithelial factors within the foregut and arcade cells.
FoxA factors may regulate a conserved epithelial program
In mammals, FoxA factors promote epithelial organ development, and their loss often correlates with EMT of tumors. We asked whether FoxA factors target core epithelial factors in mammals as they do in C. elegans. We queried existing FoxA ChIP-seq data sets for mouse cells (Soccio et al., 2011) and human cells (Motallebipour et al., 2009; Hurtado et al., 2011; Gosalia et al., 2015; Wang et al., 2015) to determine whether FoxA factors bound to the regulatory elements of epithelial factors, examining 1000 base pairs up and downstream of the gene body, as well as the gene body itself. We did not analyze enhancers, as they are often difficult to assign to a particular gene.
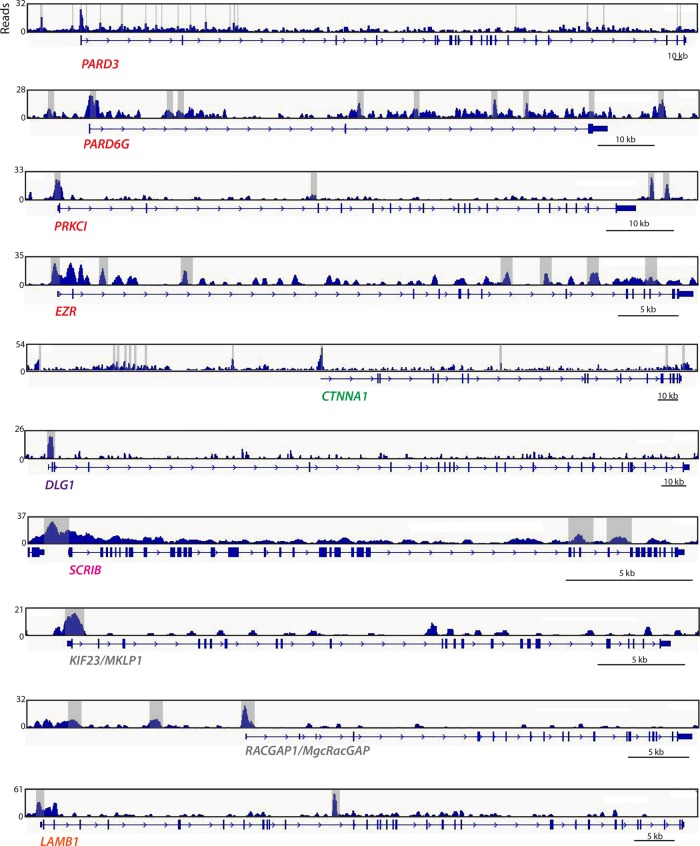
We examined homologues of the 50 epithelial-specific factors that were bound by PHA-4 in C. elegans (Table 1). We found that 27 of 50 mouse epithelial homologues had an associated FoxA2 peak in primary liver cells (Table 2). Caco2 cells are derived from colorectal cancer but mimic normal polarized enterocytes when cultured to confluency (Fogh et al., 1977; Alvarez-Hernandez et al., 1991; Meunier et al., 1995). Analysis of FoxA2 ChIP-seq data revealed FoxA2 peaks near the human homologues of 33 of 50 C. elegans epithelial genes (Gosalia et al., 2015). Intriguingly, PARD3 and PARD3B—homologues of PAR-3, a master regulator of epithelial polarity—had 11 and 13 peaks, respectively, throughout the gene body, mostly in introns, suggesting potential regulation by FoxA2. PARD3 and PARD3B were also bound by FoxA1 and FoxA2 in human embryonic stem cell–derived gut tube (GT) and foregut (FG) cells (Wang et al., 2015). In GT and FG cells, respectively, FoxA1 bound 33 and 32 of 50 worm epithelial genes, whereas FoxA2 bound 43 and 45 of 50 genes (Figure 6 and Table 2). In addition to the par-3 homologues listed here, mouse and human homologues of dlg-1 and erm-1 were also bound by FoxA1/2 in all data sets analyzed.
TABLE 2:
List of conserved epithelial factors with bound mammalian FoxA.
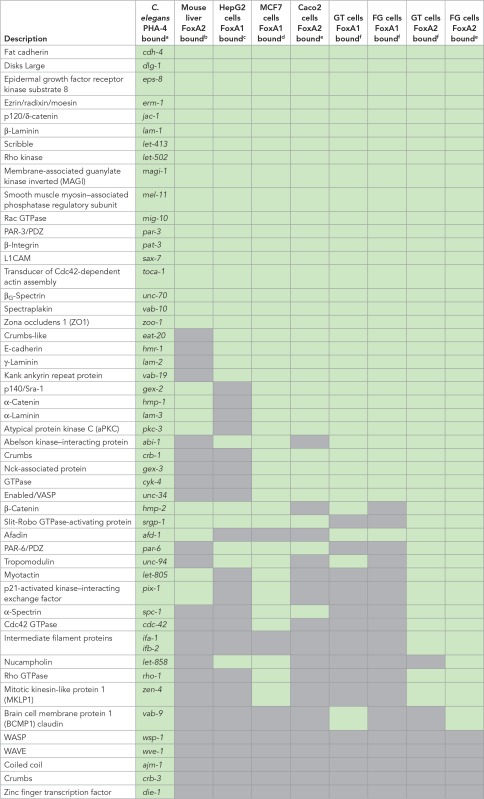 |
Green indicates that the PHA-4 or FOXA homologue binds to regulatory regions in the listed worm gene and its homologues in the listed mammalian ChIP-seq data sets. Gray boxes denote no binding for any homologues. For a complete list of the bound homologues, see Supplemental Table S2.
FIGURE 6:
FOXA2 binds to promoter and introns of human epithelial genes. Genome browser displays of FOXA2-binding peaks for selected genes from the ChIP-seq data set generated by Wang et al. (2015) from foregut-induced human embryonic stem cells. Significant FOXA2 peaks (MACS2; see Materials and Methods) are highlighted in gray. Only a single representative gene model is shown, although several have alternative variants.
We also analyzed two data sets from human cancer cells derived from epithelial organs. HepG2 cells are derived from a hepatocellular cancer (Knowles et al., 1980). FoxA1 binding was detected in 28 of 50 human homologues of nematode genes (Motallebipour et al., 2009; Table 2). Of these, 16 were shared between mouse liver and human liver cancer cells. We also examined MCF7 breast cancer cells and observed that FoxA1 bound 42 of 50 homologues of nematode epithelial genes (Hurtado et al., 2011; Table 2). In MCF7 cells, members of all three polarity groups were bound (CRB1-2, PARD3/6, DLG1-3), as were mediators of cell–cell (e.g., CDH1) and cell–matrix (e.g., ITGB1-8) interactions.
In sum, of 50 gene families surveyed, FoxA1 or FoxA2 was observed in at least seven of eight data sets for 26 of the gene families (Table 2 and Supplemental Table S2). Included in this list are genes essential for proper epithelial polarity in mammalian cells (E-cadherin, β-integrin, Discs Large), as well as mediators of tight and adherens junction formation and function (Par3, ZO1, MAGI2, α-catenin, p120-catenin, Rho kinase). These results implicate mammalian FoxA for regulation of a common set of epithelial factors in many tissue types and suggest that FoxA factors may function as master regulators of epithelial identity in multiple species.
ZEN-4 is required for protein accumulation in the arcade cell epithelium
Once transcription is activated throughout the digestive tract, the arcade cells delay accumulation of DLG-1 protein until polarity onset (Figure 2, comma stage). The ZEN-4 kinesin is essential for arcade cell polarity (Portereiko et al., 2004), and we tested whether zen-4 had a role in dlg-1 accumulation. zen-4 mutants had abundant dlg-1 mRNA at the comma/1.25-fold stages (when the arcade cells normally polarize), similar to wild-type siblings (Figure 7, A and B). However, zen-4 mutants lacked DLG-1 protein at the 1.25-fold stage (compare Figure 7, G to H). Maternally supplied par-6 and erm-1 gave similar results to dlg-1. Levels of par-6 and erm-1 RNA were virtually identical in zen-4 mutant arcade cells versus control arcade cells (Figure 7, D and F), but there was no detectable protein (Figure 7, J and L). Protein was also missing for apical (PKC-3, PAR-3) and junctional (AJM-1, HMR-1) markers, as determined previously (Portereiko et al., 2004). Using an overexpressed AJM-1::GFP reporter that increased levels in both wild-type and mutant embryos, we estimate that zen-4 mutants have 20-fold lower AJM-1 than the wild type (Supplemental Figure S4). We conclude that, normally, ZEN-4 promotes accumulation of polarity proteins to form the arcade cell epithelium.
FIGURE 7:
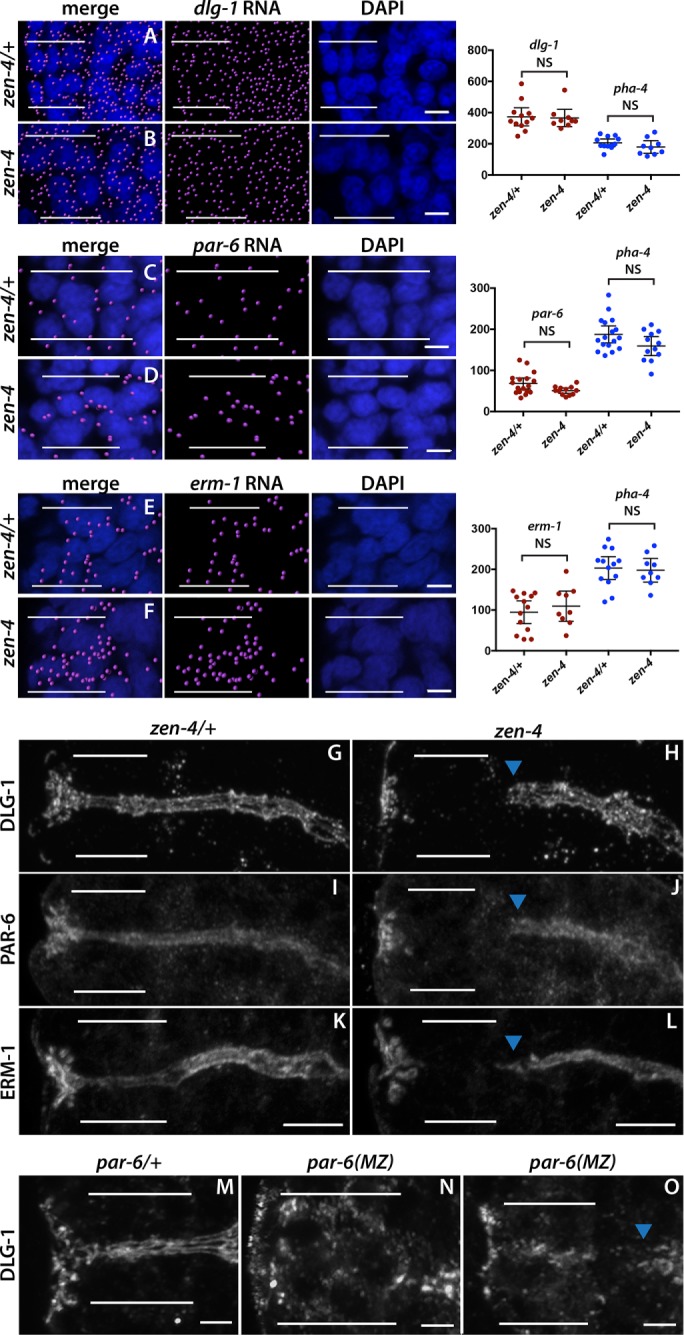
zen-4 and par-6 are required for arcade cell polarization. (A–F) mRNA detected by smFISH was imported into Imaris 3D software and is pseudocolored magenta (Materials and Methods); DAPI is blue. Epithelial factor mRNAs (dlg-1, par-6, erm-1) are present in both zen-4/+ (A, C, E) and zen-4 mutant (B, D, F) arcade cells, which are highlighted by white bars. As shown on the graphs on the right, no significant difference in the amount of RNA between zen-4/+ and zen-4 mutant arcade cells was detected (p > 0.05 for all comparisons between control and mutant; see Materials and Methods for actual values). pha-4 RNA was used as a negative control and to help localize the arcade cells for quantification and was also unchanged between zen-4/+ and zen-4 arcade cells (p > 0.05, see Materials and Methods for actual values). (G–L) PAR-6 (G), DLG-1 (I), and ERM-1 (K) are expressed and localized to the apical/junctional domain in zen-4/+ arcade cells (white bars). These proteins are not detected in zen-4 mutant arcade cells (H, J, L) but are expressed and localized in the neighboring foregut (arrowhead). (M–O) DLG-1 is expressed and localized in the arcade cells (white bars) of par-6/+ (M) embryos. Forty percent (14 of 35) of par-6 mutant embryos contain DLG-1, but it is mislocalized (N). In the other 60% (21of 35), DLG-1 is properly localized but fails to form continuous junctions, similar to the adjacent foregut (blue arrowhead; O). All images are maximum intensity projections. Anterior is to the left. Scale bars, 3 μm (A–F), 5 μm (G–L), 2 μm (M–O).
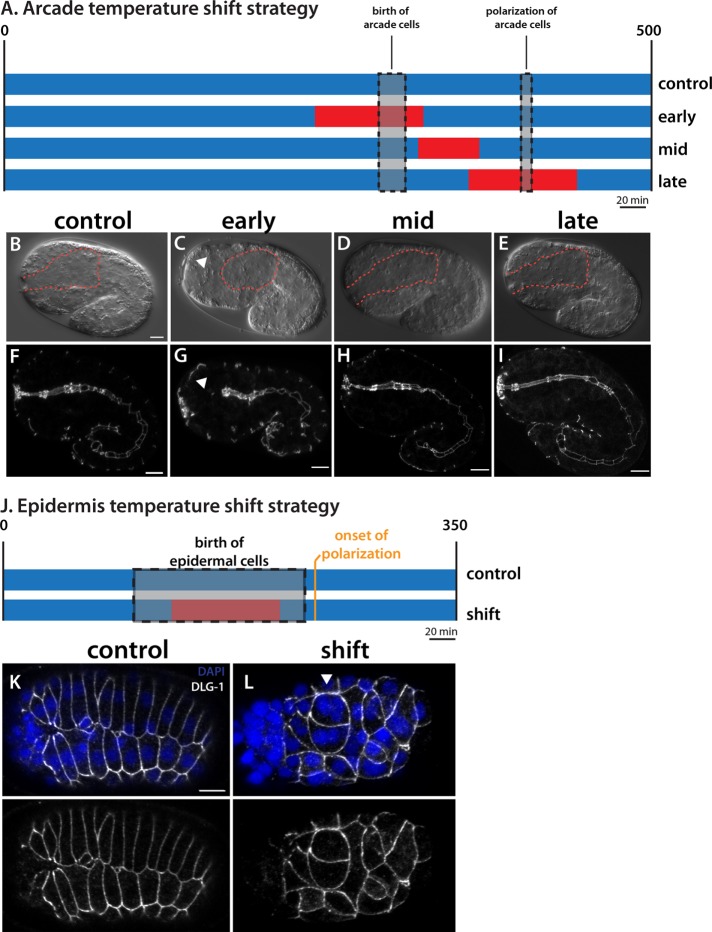
The effect of zen-4 on accumulation of polarity proteins suggested that ZEN-4 would function early, before polarity onset. To test this idea, we performed temperature-shift experiments to pinpoint when zen-4 activity was required (Figure 8). We used a temperature-sensitive allele of zen-4, or153ts (Severson et al., 2000), which abrogates binding to its obligate partner in cytokinesis, cyk-4, when shifted to the restrictive temperature (Pavicic-Kaltenbrunner et al., 2007). We inactivated zen-4 during three time periods: close to the time of arcade cell birth (early); during arcade cell polarization (late); and between the early and late time points (middle). Embryos were shifted to restrictive temperature for a 70-min window, followed by incubation at permissive temperature to the 1.5-fold stage, when the arcade cells have formed an epithelium in the wild type. We confirmed rapid inactivation of zen-4 at these late stages by testing for cytokinesis defects (Supplemental Figure S5), which is a canonical phenotype of or153ts (Severson et al., 2000).
FIGURE 8:
ZEN-4 activity is required at arcade cell birth. (A) The temperature-shift strategy. Rectangles denote developmental time (scaled to 20°C). Blue represents permissive temperature (15°C), and red denotes restrictive temperature (26°C). The control sample was incubated at permissive temperature only. The early shift started ∼50 min before arcade cell birth, the late shift started ∼50 min before arcade cell polarization, and the mid shift is in between. Representative DIC (B–E) and DLG-1 (F–I) images of embryos after the foregoing incubations. The foregut-associated basement membrane (BM) is highlighted by a red dotted line. The control, mid, and late embryos all have a polarized arcade cell epithelium spanning the epidermis and foregut. Note that the BM surrounds an elongated foregut. The early shift results in an unattached foregut because the arcade cells (arrowhead) failed to polarize (C, G). Note that the BM completely surrounds the foregut primordium, which is unattached to the anterior epidermis (C), and that DLG-1 is not expressed in the arcade cells (G). (J) The epidermal temperature-shift strategy. The epidermal cells are born over a longer period of time than the arcade cells (gray), and this period encompasses the time at the restrictive temperature (red). Embryos were shifted back to permissive temperature before the onset of epidermal polarization. (K, L) Representative maximum intensity projections through the epidermis of embryos at the end of the shift. In the control embryo (K), DLG-1 (white) is found at the cell junctions of epidermal cells that have formed two rows on the dorsal surface. These cells are mononucleate. In the shifted embryo (L), the epidermal cells generate and localize DLG-1 properly, even though many cells are obviously multinucleate (arrowhead) due to cytokinesis defects. Anterior is to the left. Scale bars, 5 μm.
The majority of embryos shifted to nonpermissive temperature at the time of arcade cell birth had defects in polarization (84%; 27 of 32; Figure 8). On the other hand, almost all embryos shifted during the middle and late periods appeared wild type (35 of 37 and 55 of 56 for the middle and late windows, respectively; Figure 8). These data suggest that ZEN-4 is required around the time of arcade cell birth rather than at the time of polarization, consistent with the idea that ZEN-4 acts early to produce polarity proteins, which are later assembled into apical and basolateral domains. As a control, inactivation of zen-4(ts) at the birth of the epidermal cells did not disrupt polarity in the epidermis, indicating that zen-4 is required specifically in the arcade cells and that blocked cytokinesis is not sufficient to disrupt epithelial polarity (Figure 8).
Structure–function analysis of ZEN-4 reveals a requirement for binding to CYK-4/GAP
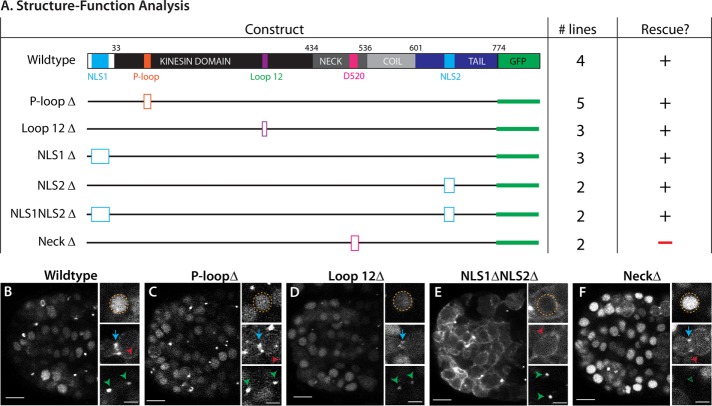
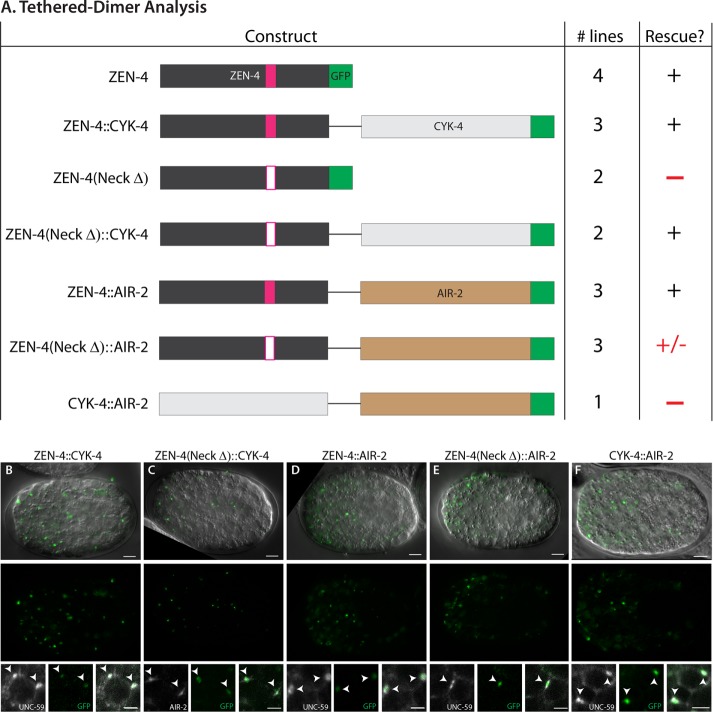
ZEN-4/MKLP relies on four protein domains to fulfill its roles in cytokinesis and neurite outgrowth (White and Glotzer, 2012; del Castillo et al., 2015). We performed structure–function analysis to gain insight into which domains were important for arcade cell polarization (Figure 9A). We assayed both rescue of the zen-4(px47) mutant phenotype (see Materials and Methods) and localization of a GFP-tagged reporter. To monitor px47 rescue, we examined progeny from heterozygous zen-4(px47) mothers. Normally 25% offspring are zen-4(px47) homozygotes, and of these, half or more arrest with unpolarized arcade cells (Table 3 and Supplemental Table S3). A wild-type ZEN-4::GFP transgene rescued homozygotes, producing ∼0% embryonic lethality from heterozygous mothers (Figure 9A, Table 3, and Supplemental Table S3). These animals displayed a dynamic localization pattern for ZEN-4::GFP that mimicked endogenous ZEN-4 and its homologues (Figure 9B; Powers et al., 1998; Raich et al., 1998; Deavours and Walker, 1999; Chen et al., 2002; Minestrini et al., 2002, 2003; Verbrugghe and White, 2007). ZEN-4::GFP was nuclear in interphase cells, localized to the ingressing furrow and cortex during mitosis, and remained at the division remnant after abcission.
FIGURE 9:
ZEN-4 structure–function analysis reveals the importance of CYK-4/GAP binding. (A) The domain structure of wild-type ZEN-4 (top). There are two nuclear localization sequences (NLS; light blue) and the globular tail domain (dark blue). The kinesin domain (black) contains the known ATP-binding pocket (P-loop, orange) and loop 12 for binding microtubules (purple). The neck domain (medium gray) contains residues important for binding to CYK-4/GAP, including D520. There is also a coiled-coil domain (light gray). GFP is linked to the C-terminus (green). To test the function of each domain in polarity, we generated small deletions (open boxes) to remove key residues for each domain. The number of independent transgenic lines is given (# lines). A plus sign designates rescue of zen-4(px47) arcade cell polarity, whereas a minus sign denotes lack of rescue. See Table 3 and Supplemental Table S3 for more details. (B–F) Live images of ZEN-4::GFP constructs at the 16E stage. Large overviews show the embryo anterior. Insets highlight nuclei (orange), furrow (blue arrow), cortex (red), or division remnants (green) from the same embryo but for different focal planes. (B) Wild-type ZEN-4::GFP. (C, D) Kinesin mutants (P-loopΔ, Loop 12Δ) have wild-type localization. (E) The double NLS deletion (NLS1ΔNLS2Δ) is reduced in the nucleus. (F) The NeckΔ is enriched in the nucleus and at the furrow/cortex, but localization to the division remnant is lost (green open arrowhead). Anterior is left. Scale bars, 5 μm (overviews), 2 μm (close-ups).
TABLE 3:
ZEN-4 structure–function analysis.
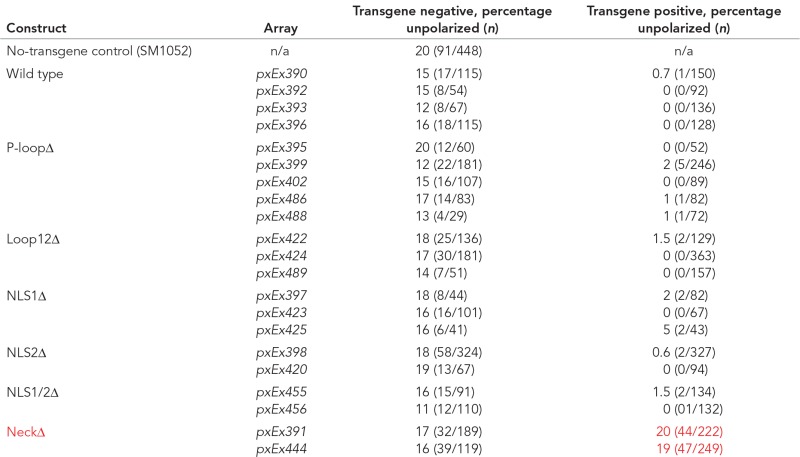 |
The left column displays the ZEN-4 structure/function construct expressed in the listed transgenic lines. SM1052 is the reference strain with genotype zen-4 dpy-20/bli-6 unc-24, maintained as a heterozygote. The progeny, by Mendelian genetics, should be 75% wild type and 25% mutant, of which the majority displays the unpolarized arcade cell phenotype (assessed by Nomarski optics, looking for foregut attachment, which occurs via arcade cell polarization), whereas a subset display a failure in epidermal enclosure and subsequently elongation. To assess for rescue, compare the percentage unpolarized between the transgene-negative (control) and transgene-positive columns for a given transgenic line. For example, pxEx390 displays 15% unpolarized arcades for the transgene-negative embryos, whereas only 0.7% of the transgene-positive embryos have unpolarized arcades and thus are considered to rescue the phenotype. By these criteria, only the NeckΔ construct fails to rescue the arcade cell polarity defect of zen-4(px47) mutants (compare red text to black text in the same column). The n for a given phenotype in a given transgenic line is in parentheses.
ZEN-4(NeckΔ)::GFP lacks five residues around residue D520 within the neck domain. This construct was unable to rescue zen-4(px47): zero of two transgenic ZEN-4(NeckΔ)::GFP lines rescued zen-4(px47) mutants, like the no-transgene control (Figure 9A, Table 3, and Supplemental Table S3). The neck domain serves two purposes. It is essential to bind CYK-4/MgcRacGAP, a GTPase-activating protein that controls ZEN-4 kinesin activity (Mishima et al., 2002; Pavicic-Kaltenbrunner et al., 2007; White et al., 2013). It is also important to localize ZEN-4 to the cell cortex and division remnant (Figure 9F). To determine whether ZEN-4 requires CYK-4 activity or division remnant localization, we tethered ZEN-4(NeckΔ) to CYK-4 using a flexible linker (see Materials and Methods; Neuhold and Wold, 1993; Castanon et al., 2001). As a positive control, we linked CYK-4 to wild-type ZEN-4. As shown in Figure 10A, Table 4, and Supplemental Table S4, both wild-type and mutant fusions were sufficient for rescue, indicating that association with CYK-4 is critical for ZEN-4 function and disrupted by the NeckΔ mutation.
FIGURE 10:
ZEN-4 tethered dimer analysis demonstrates the importance of the ZEN-4/CYK-4 interaction. (A) Experimental design. Wild-type ZEN-4 or ZEN-4(NeckΔ) were tethered to either GFP-tagged CYK-4 or AIR-2 (AuroraB kinase) via a flexible linker (black line). CYK-4 was tethered to GFP-tagged AIR-2 (bottom construct; note that additional lines were generated, and these did not express or rescue; unpublished data). The number of lines and ability to rescue the zen-4(px47) arcade cell polarization defect are shown. The untethered ZEN-4 or the ZEN-4(NeckΔ) data from Figure 9 are shown for reference. See Table 4 and Supplemental Table S4 for more details. (B–F) Live imaging of GFP-tagged ZEN-4 tether constructs at the 16E stage. Single focal plane through the digestive tract. Top, a merge of DIC and tether::GFP (green), which is also displayed alone in the middle row. Most signal (green) is found at division remnants (white arrowheads, bottom) by colocalization with known division remnant markers UNC-59/septin (Nguyen et al., 2000) or AIR-2 (Schumacher et al., 1998) (both white). UNC-59 is enriched at the division remnant after furrow ingression. Anterior is left. Scale bars, 5 μm (whole embryo), 2 μm (close-ups).
TABLE 4:
Tethered dimer analysis.
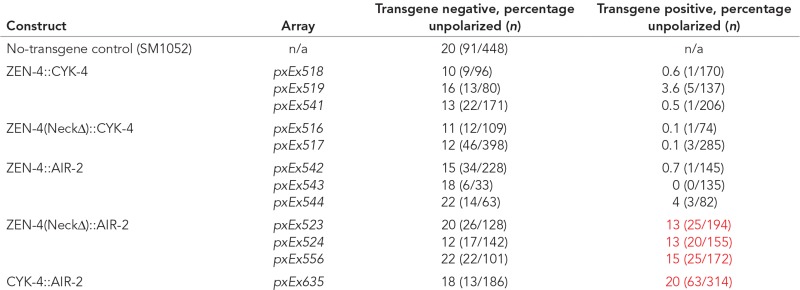 |
The left column displays the tether construct in the listed transgenic lines (Array). SM1052 is the reference strain with genotype zen-4 dpy-20/bli-6 unc-24, maintained as a heterozygote. The progeny should be 75% wild type and 25% mutant, of which the majority displays the unpolarized arcade cell phenotype. To assess for rescue, compare the percentage unpolarized between the transgene-negative (control) and transgene-positive columns for a given transgenic line. A comparison of transgene-negative to transgene-positive progeny reveals that restoring CYK-4 binding to ZEN-4(NeckΔ) is able to rescue the arcade cell polarity defect of zen-4(px47) mutants, whereas binding to AIR-2 may partially rescue. Restoring CYK-4 to the division remnant in the absence of ZEN-4 via tethering to AIR-2 is unable to rescue, demonstrated by the quantified line and the 13 additional lines containing either wild-type or kinase-dead AIR-2 (unpublished data). Polarity defects were scored by Nomarski optics, assessing whether the foregut had attached to the epidermis, which occurs via arcade cell polarization. Red lettering is indicative of a failure to fully rescue (compare to black text within the same column).
The tethered dimer localized ZEN-4::CYK-4 to the division remnant (Figure 10, B and C). To determine whether localization was sufficient for rescue, we tethered ZEN-4 to the Aurora kinase AIR-2/AuroraB, which is enriched at the division remnant partially overlapping with CYK-4 (Schumacher et al., 1998) and is required to localize ZEN-4 normally (Severson et al., 2000). A wild-type ZEN-4::AIR-2 fusion served as a control. AIR-2 was only weakly able to rescue ZEN-4(NeckΔ) despite complete localization to the division remnant (Figure 10, Table 4, and Supplemental Table S4). Embryos bearing ZEN-4(NeckΔ)::AIR-2 had 18% lethality (range 17–19%), compared with ∼23% lethality for no-transgene control embryos (range 17–27%). The wild-type ZEN-4::AIR-2 control was fully functional (0–4% lethality for array+ embryos), demonstrating that AIR-2 did not interfere with ZEN-4 function. We next queried whether the function of ZEN-4 was simply to ensure that CYK-4 traffics to the division remnant. To test this possibility, we tethered CYK-4 to AIR-2. Although the CYK-4::AIR-2 tether was indeed localized to the division remnant (Figure 10F), it was unable to rescue zen-4(px47) mutants (∼20% of both transgene+ and no-transgene control embryos displayed polarity defects; Table 4 and Supplemental Table S4). We conclude that accumulation at the division remnant is not sufficient for rescue and that both CYK-4 and ZEN-4 are needed for polarization.
CYK-4 activates the ATPase and kinesin activities of ZEN-4 on microtubules (Mishima et al., 2002; White et al., 2013; Tao et al., 2016). To determine whether microtubules were critical for ZEN-4 function in polarity, we mutated the P-loop and loop 12 within the kinesin domain (Figure 9A). The P-loop contains the ATPase activity of the kinesin (Saraste et al., 1990; Nakata and Hirokawa, 1995), and mutations in this domain in ZEN-4 homologues result in a motor-dead protein that can still bind microtubules (Chen et al., 2002; Matuliene and Kuriyama, 2002). Loop 12 of kinesins is important for binding to microtubules (Woehlke et al., 1997; Raich et al., 1998). These mutations had a mild defect on ZEN-4 localization and did not affect rescue (Figure 9, C and D, Table 3, and Supplemental Table S3). Thus we have no evidence that microtubule activities are important for ZEN-4 during polarity. Finally, we note that ZEN-4 normally localizes to nuclei during interphase. Disruption of two predicted nuclear localization signals (NLSs) strongly reduced nuclear accumulation but had no observable effect on rescue (Figure 9E, Table 4, and Supplemental Table S4). Similarly, mutation of either NLS1 or NLS2 alone did not affect ZEN-4 activity (Figure 9A, Table 3, and Supplemental Table S3). These data suggest that neither the kinesin function nor nuclear localization is necessary for polarity, although we cannot rule out that residual ZEN-4 in the nucleus or movement on microtubules is sufficient for rescue. In addition, nuclear localization is unperturbed in the NeckΔ mutant, which suggests that nuclear localization may not be sufficient for polarity.
CYK-4 can modulate actin dynamics by way of its GTPase-activating domain (Jenkins et al., 2006; Canman et al., 2008; Loria et al., 2012; Zhang and Glotzer, 2015; Zhuravlev et al., 2017). We previously determined that actin is severely disrupted in zen-4(px47) mutant arcade cells (Portereiko et al., 2004). Therefore we determined whether a gain-of-function allele of the RhoGEF ect-2, which suppresses the actomyosin-driven cytokinetic furrow ingression defect of cyk-4 mutants (Zhang and Glotzer, 2015), could also rescue the polarity defect of zen-4 mutant arcade cells. We observed no suppression, as ect-2(xs110); zen-4(px47) double mutants had a similar percentage of embryos with unpolarized arcade cells as the zen-4(px47) control (17 and 16%, respectively; Supplemental Table S5). These data do not rule out a role for actin dynamics, however, as the ect-2(gf) allele may not be strong enough to overcome the zen-4 polarity defect. In summary, ZEN-4 relies on its interaction with CYK-4 to induce polarity. On the other hand, the microtubule-related ATPase and kinesin activities of ZEN-4 are dispensable for epithelium formation.
PAR-6 is required to position DLG-1 within nascent arcade cell epithelia
The data suggest that PHA-4 and ZEN-4 control RNA and protein production of polarity factors in the arcade cells. Once produced, standard polarity pathways likely organize the apical, junctional, and basolateral domains within the arcade cells (Rodriguez-Boulan and Macara, 2014). We tested this idea by examining par-6 mutants. Because par-6 is important for the first embryonic cell divisions, we removed PAR-6 protein at the gastrula stage, using an elegant ZF1 system (par-6m+z-; Nance et al., 2003; Totong et al., 2007). par-6 mutants produced DLG-1 protein (Figure 7, N and O) but often failed to localize it properly to the junctional domains. Instead, we detected DLG-1 in the cytoplasm of par-6(m+z-) mutants (Figure 7N). These data reveal that temporal regulation of the arcade cell epithelium depends on three levels of tissue-specific control: first, at the transcriptional level; second, at the level of protein expression; and third, at the level of protein localization to nascent adherens junctions.
DISCUSSION
The past three decades have seen major advances in our understanding of cell polarization during MET (Chaffer et al., 2007). Little is known, however, about coordination between distinct epithelia to produce the mature body plan. Our data show that C. elegans epithelia polarize just before the morphogenetic event in which that epithelium is involved (e.g., epiboly, torus formation). In particular, the arcade cells delay polarization until after the epidermis and gut epithelia have matured sufficiently to withstand the pulling forces of embryo elongation. In support of this notion, if the arcade cells polarize before the epidermis is fully mature (e.g., as in fbn-1/fibrillin mutants), the epidermis is pulled into the embryo by pulling forces from the digestive tract, likely due to resistance of the foregut to stretching or lengthening (Kelley et al., 2015). We discovered four regulators that ensure delayed timing in arcade cells: PHA-4/FoxA initiates expression of polarity genes within the arcade cells, ZEN-4/MKLP1 and CYK-4/MgcRacGAP dictate the onset of protein accumulation, and PAR-6 controls subcellular localization within the nascent epithelium.
PHA-4/FoxA regulates a complete epithelial program
The FoxA pioneer transcription factor PHA-4 promotes the onset of dlg-1 expression within the foregut and arcade cells but not other cell types. In addition to dlg-1, the majority of apical, junctional, and basolateral genes had an associated PHA-4 ChIP peak (Figure 5 and Table 1); most associated either with the TSS or within the first intron, which are hallmarks of bona fide PHA-4 target genes (Gaudet and Mango, 2002; Gaudet et al., 2004). All of these genes are also expressed in other epithelia (or other cell types). We confirmed that expression for two of these factors (PAR-3, ERM-1) was selectively reduced in the foregut and arcade cells (Figure 3). This rules out the possibility of a single epithelial program regulated by a common set of transcription factors in all epithelial organs and suggests that the mechanism of timing is controlled at the tissue and organ levels by tissue/organ-specific transcription factors such as PHA-4.
The P4.9dlg-1 construct was able to recapitulate the endogenous expression of DLG-1 protein in the foregut/arcade cells and the epidermis (Figure 4). A study of the C. elegans transcriptional start site and enhancer landscape identified a putative enhancer for dlg-1 at ∼6 kb upstream of the ATG start (Chen et al., 2013). Our analysis revealed that this enhancer was not necessary for normal dlg-1 expression in the foregut/arcade cells, at least in the context of a complex multicopy array. It is possible that this region functions as a shadow enhancer that promotes robust dlg-1 expression but is not essential (Hong et al., 2008; Perry et al., 2010). In addition, the P4.9dlg-1 construct carried a heterologous 3′ untranslated region (UTR), suggesting that the endogenous 3′UTR is not necessary for proper expression in the foregut and arcade cells. Perhaps regulation occurs through the 5′UTR, as suggested for some posttranscriptional events in C. elegans (Lee and Schedl, 2004; Jungkamp et al., 2011; Vora et al., 2013). Note that in the midgut, some cells expressed P4.9dlg-1 RNA one cell cycle earlier (unpublished data), suggesting additional cis-regulatory sites for this tissue.
FoxA regulation of a general epithelial program may be conserved
We extended our analysis to two mammalian homologues of PHA-4: FoxA1 and FoxA2. FoxA factors are necessary for the proper development of many epithelial organs, mostly of endodermal origin (e.g., liver, pancreas, intestine, lung). Conversely, epithelial tumors often lose FoxA expression during EMT (Tang et al., 2011; Wang et al., 2014; Bersaas et al., 2016). These studies suggested that FoxA acts by suppressing the transcription factor Slug, a known regulator of EMT, other transcription factors, and adhesion signaling pathways (Song et al., 2010; Tang et al., 2011; Wang et al., 2014). Our survey revealed FoxA1/FoxA2 binding to the promoter and introns of mammalian homologues of the 50 epithelial-specific worm genes with PHA-4 binding (Figure 5, Table 2, and Supplemental Table S1), suggesting that FoxA1/A2 may promote a general epithelial program in mammals as in worms. We note that work on the Drosophila trachea also demonstrates that the combined efforts of a cell-fate regulator and its effectors are necessary for proper epithelial morphogenesis (Brodu and Casanova, 2006; Shaye et al., 2008), suggesting that the strategy we described for PHA-4 and its effectors is conserved and used in mammals as well.
ZEN-4 and PAR-6 control polarity onset
Our analysis revealed a significant delay between the appearance of dlg-1 transcripts and the accumulation of protein within the arcade cells as compared with the other epithelia. ZEN-4 was critical for this regulation, as transcripts were abundant in zen-4 mutant arcade cells, but protein was absent for three of three genes tested (Figure 7). We note that absence of protein was not due to mislocalization within cells, because par-6 mutants displayed bountiful mislocalized DLG-1 (Figure 7) and ERM-1 (Von Stetina and Mango, 2015). Instead, ZEN-4 likely regulates the translation or stability of DLG-1 and other polarity proteins. This role suggests that ZEN-4 acts upstream of PAR-6, which is in agreement with the absence of PAR-6 in zen-4 mutants and the temperature-sensitive period of zen-4(ts) mutants. These data extend our previous finding that the arcade cells are properly specified in zen-4 mutants (Portereiko et al., 2004).
ZEN-4 is a member of the kinesin-6 family and possesses multiple structural domains. One study suggests that a kinesin-6 member may traffic ribonucleoprotein stress granules to promote localized translation (Taniuchi et al., 2014). However, this is unlikely to be how ZEN-4 regulates polarity protein production, as our data demonstrated that kinesin activity and microtubule binding were not necessary. Our structure–function analysis also showed that nuclear localization was dispensable for ZEN-4 function, but binding to CYK-4/GAP was essential (Figure 9, Table 3, and Supplemental Table S3). These data suggest that regulation of actin rather than microtubules may be the critical parameter for polarization. Consistent with this idea, actin, but not tubulin, is dramatically disrupted in zen-4 mutant arcade cells (Portereiko et al., 2004; unpublished data). Previous studies revealed that ZEN-4 and CYK-4 can modulate actin dynamics by regulating RhoA and Rac activity (Jenkins et al., 2006; Canman et al., 2008; Loria et al., 2012; Zhang and Glotzer, 2015; Zhuravlev et al., 2017), and therefore they could be functioning in a similar manner in polarity. Another possibility is that ZEN-4 and CYK-4 act as a scaffold to promote accumulation of polarity proteins at the appropriate time.
Tethering of mutant ZEN-4 to the division remnant gave either full (CYK-4) or partial (AIR-2) rescue (Figure 10, Table 4, and Supplemental Table S4). This result raises the intriguing possibility that localization to the remnant may contribute to ZEN-4 function. The division remnant has been implicated in other polarizing events, such as positioning of dendrites in neuronal cells and establishing the dorsoventral axis in C. elegans (Pollarolo et al., 2011; Singh and Pohl, 2014; Dionne et al., 2015). Marking a site between dividing cells has been studied most extensively in yeast, where the bud scar serves as a marker for the next division (reviewed in Casamayor and Snyder, 2002; Slaughter et al., 2009; Bi et al., 2012).
The data presented here offer a possible explanation for the selectivity of zen-4 for the arcade cells. Only the arcade cell epithelium is disrupted in zen-4 mutants; other epithelia polarize normally (Portereiko et al., 2004; Hardin et al., 2008). Wild-type arcade cells exhibited ∼100-min delay between the onset of dlg-1 RNA and protein, whereas other epithelia displayed no or a much shorter delay. An appealing hypothesis is that the arcade cells depend on an extra layer of posttranscriptional regulation, mediated by ZEN-4 and CYK-4, compared with other embryonic epithelia. This extra layer of regulation may be critical to ensure that polarization occurs late in embryogenesis.
Homologues of ZEN-4 and CYK-4 have been implicated in polarity at later stages of epithelium formation. These factors modulate RhoA and Rac1 in epithelial junctions of vertebrate cells (Ratheesh et al., 2012; Guillemot et al., 2014; Breznau et al., 2015). This activity may be missing or redundant in C. elegans because conditional inactivation of zen-4 did not reveal a requirement at mid or late time points. The present evidence from the ect-2(gf) epistasis test does not definitively support or refute a role for RhoA/Rac1 regulation during establishment of polarity in C. elegans epithelia (Supplemental Table S5). Note that the vertebrate studies were not designed to study a possible early role in polarity protein accumulation, so it remains to be seen whether ZEN-4 function is conserved.
Once DLG-1 and other polarity proteins are produced, they are rapidly localized within the cell. We found that par-6 was necessary for localization of DLG-1 within the arcade cells in ∼50% of tested embryos (Figure 7). These data extend our previous observations that in half of the tested embryos, several markers of epithelial polarity (actin, HMR-1/cadherin, ERM-1, AJM-1) failed to localize properly within the arcade cells in par-6 mutants (Von Stetina and Mango, 2015). In the other half, protein is localized properly (like DLG-1) but fails to form continuous junctions between other arcade cells and between tissues (i.e., foregut and arcades). In other epithelia, this failure to mature junctions is the most severe par-6 phenotype observed, but cell polarity is intact (Totong et al., 2007; Von Stetina and Mango, 2015). What accounts for the difference? One possibility is that distinct tissues are differentially dependent on specific proteins. For example, PAR-6 is more important in the epidermis than PAR-3, whereas par-3 mutants have a more severe phenotype in the midgut than par-6 mutants (Achilleos et al., 2010). By this logic, PAR-6 may be a more important player for polarization in arcade cells compared with other tissues. An alternative possibility is that residual PAR-6 may provide polarizing activities. The M/Z system removes wild-type protein expressed from maternally deposited transcripts, but the system takes time to complete. Perhaps residual PAR-6 is present and active when other epithelia are formed but gone by the time the arcade cells undergo polarization. Consistent with this idea, embryos with partial arcade cell polarity always exhibited some PAR-6 protein, whereas those that completely lacked detectable PAR-6 did not (Von Stetina and Mango, 2015).
MATERIALS AND METHODS
Strains and maintenance
Nematodes were grown at 20°C using standard conditions (Brenner, 1974), unless otherwise stated. Some strains were provided by the Caenorhabditis Genetics Center. Strains used: N2 (SM1880); SM469 [pxIs6 (pha-4::GFP::Histone)] (Portereiko and Mango, 2001); SM1052 [zen-4 (px47) dpy-20 (e1282)/bli-6(sc16) unc-24(e138) IV] (Portereiko et al., 2004); SM1271 [zen-4 (px47) jcIs1 (AJM-1::GFP, pRF4)/bli-6(sc16) unc-24(e138) IV] (Portereiko et al., 2004); SU93 [jcIs1 (AJM-1::GFP, pRF4)] (Koppen et al., 2001); FZ223 [mcIs204 (P7dlg-1::DLG-1::GFP; pRF4)] (McMahon et al., 2001); JJ1743 [par-6(tm1425)/hIn1[unc-54(h1040)] I; him-8(e1489) IV] (Totong et al., 2007); FT36 [unc-101(m1) par-6(zu170)I; zuIs43 (pie-1::GFP::PAR-6::ZF1 + unc-119(+))] (Totong et al., 2007); and EU554 [zen-4(or153) IV] (Severson et al., 2000). For a description of the par-6MZ genetic strategy, see Von Stetina and Mango (2015). Strains generated for this study are listed in Supplemental Table S6.
Temperature-shift experiments
zen-4(or153ts) was maintained at 15°C. To perform arcade cell temperature shifts on staged embryos, gravid adults were transferred to a precooled (4°C) dissection chamber with 50 μl of ice-cold M9 Buffer (22 mM KH2PO4, 22 mM Na2HPO4, 85 mM NaCl, 1 mM MgSO4). Embryos were released by dissecting the adults using 26-gauge needles. Two-cell embryos were quickly collected and transferred via mouth pipette to a 0.2-ml PCR tube or to a polylysine-coated slide on ice. Once 10–30 two-cell embryos were collected, the tube was placed in a Bio-Rad PCR machine precooled to 15°C or the slide was placed into a humidified chamber at 15°C. Embryos were incubated at 15°C until the appropriate stage was reached and then shifted to 26°C to inactivate zen-4. After the inactivation period was over, embryos were shifted back to 15°C until the 1.5- to 2-fold stage, when embryos from tubes were transferred to a 4% agar pad on a glass slide to assess foregut attachment by differential interference contrast (DIC) microscopy or the slides were processed for immunostaining (see later description). Incubation times were determined based on Sulston's embryonic lineage timing at 20°C and adjusted accordingly; 15°C was considered 1.5 times slower than 20°C, whereas 26°C was considered 1.2 times faster. Timing for specific stages for N2 and zen-4(or153ts) was determined with test incubations (unpublished data). The specific programs for each shift were as follows:
Early shift (circa arcade cell birth): 360 min at 15°C (equivalent of 240 min at 20°C), 70 min at 26°C (equivalent of 84 min at 20°C), and 270 min at 15°C (equivalent to 180 min at 20°C).
Mid shift (between birth and polarization): 480 min at 15°C (equivalent of 320 min at 20°C), 41 min at 26°C (equivalent of 49.2 min at 20°C), and 140 min at 15°C (equivalent to 93 min at 20°C).
Late shift (circa polarization): 540 min at 15°C (equivalent of 360 min at 20°C), 70 min at 26°C (equivalent of 84 min at 20°C), and 60 min at 15°C (equivalent to 50 min at 20°C).
Temperature shifts for epidermal cells were performed in a similar manner, in which dissected two-cell embryos were transferred to a polylysine-coated slide and incubated in a humidified chamber at the appropriate temperature. Control embryos were incubated at 15°C for 525 min (equivalent of 350 min at 20°C). Shifted embryos were incubated for 195 min at 15°C (equivalent of 130 min at 20°C), 70 min at 26°C (equivalent of 84 min at 20°C), and 205 min at 15°C (equivalent to 137 min at 20°C). The slides were then processed for immunostaining.
Embryos for arcade cell shifts were scored for unattached foreguts using DIC optics on a Zeiss AxioImager M2 upright compound microscope with a 63× objective. Alternatively, embryos from the arcade cell shifts were fixed and immunostained for DLG-1 expression and imaged using a Zeiss LSM880 confocal microscope. To determine whether the canonical phenotype associated with zen-4ts was obtained, embryos were processed and incubated using the early shift settings but removed from the protocol 10 min after returning to the permissive temperature. These embryos were adhered to polylysine slides, fixed, and immunostained for ERM-1 expression and imaged via structured-illumination on a Zeiss AxioImager M2 with Apotome. Embryos for epidermal experiments were fixed, immunostained to detect DLG-1, and assessed for multinucleate cells and polarity using a Zeiss LSM880 confocal microscope.
Molecular biology
Supplemental Table S7 lists the primers used. For all PCR reactions, either AccuStart HiFi Taq Polymerase (QuantaBio) or PrimeStar Taq Polymerase (Takara) was used.
pML902 (P7dlg::DLG-1::GFP::unc-54 3′UTR; McMahon et al., 2001) was used as the template to generate P3.9dlg::DLG-1::GFP::unc-54 3′UTR (Supplemental Figure S6), P4.3dlg::DLG-1::GFP::unc-54 3′UTR, and P4.9dlg::DLG-1::GFP::unc-54 3′UTR. After amplification, a Qiagen PCR Purification kit was used to clean up the reaction. The PCR products were subsequently used for injection to generate transgenics (see later description).
ZEN-4::GFP (bsem1129) was generated by swapping out GFP for yellow fluorescent protein (YFP) from bsem1105 (ZEN-4::YFP) using NcoI and BamHI. bsem1129 contains genomic DNA from 3 kb upstream of the predicted start site, the entire coding region up to the stop codon, fused with GFP and the unc-54 3′UTR from pPD95.77 (Addgene plasmid 1495, a gift from Andy Fire, Stanford University).
ZEN-4 deletion constructs were generated by a variation of overlap PCR (Hobert, 2002), where the overlap primers contained sequence adjacent to the region to be removed and replaced with a KasI restriction site. Supplemental Table S7 lists the specific nucleotides (and the corresponding amino acids) removed in each construct. Nuclear localization sites were predicted by PSORTII (http://psort.hgc.jp/form2.html; Nakai and Horton, 1999). bsem1129 was used as the template to generate each 5′ and 3′ fragment, which were cleaned using the Qiagen PCR purification kit. We used 10 ng of each 5′ and 3′ fragment for the overlap reaction. Overlap products were gel-purified (Qiagen) and cloned into pGEM-T (Promega) or pCR4Blunt-TOPO (Invitrogen). The NLS1/NLS2 double deletion was generated by subcloning a BclI-generated fragment from bsem1231 (ZEN-4::NLS1Δ::GFP) into the BclI site of bsem1210 (ZEN-4::NLS2Δ::GFP). Constructs were verified to have the expected nucleotides removed, and no deleterious mutations, by Sanger sequencing.
CYK-4::GFP (bsem1289) was generated via overlap PCR by amplifying the Caenorhabditis briggsae CYK-4 genomic coding sequence (up to the stop codon) from bsem1093 and fusing it to GFP::unc-54 3′UTR from pPD95.77. AIR-2::GFP (bsem1292) was generated via overlap PCR by amplifying the AIR-2 genomic coding sequence (up to the stop codon) from N2 genomic DNA and fusing it to GFP::unc-54 3′UTR from pPD95.77. The resulting overlap PCR products were subcloned into pCR4-TOPOBlunt and sequence verified. Each half of the ZEN-4 tether constructs was generated by PCR, with the added tether sequence as an overhang. The flexible tether sequence chosen has successfully generated forced dimers in Drosophila (Neuhold and Wold, 1993; Castanon et al., 2001).
To generate the CYK-4::AIR-2 tether, CYK-4 with a modified tether in the 3′ primer (second codon changed to generate a KpnI site; see Supplemental Table S7 for more details) was amplified from bsem1093 and subcloned into pCR4-TOPOBlunt to create bsem1480. The tether that precedes AIR-2::GFP was also modified to the KpnI version using overlap PCR and subcloned into pCR4-TOPOBlunt to create bsem1336. The KpnI/SphI fragments bsem1480 and bsem1336 were ligated together to create bsem1481 (Pcyk-4::CbCYK-4::Kpn-tether::AIR-2::GFP_TOPO). Overlap PCR was used to remove K59 (Bishop and Schumacher, 2002; Rogers et al., 2002) and the preceding four amino acids to generate a kinase-dead version of AIR-2::GFP, which was subcloned into pCR4-TOPOBlunt to create bsem1483. The KpnI/SphI fragments bsem1481 and bsem1483 were ligated together to create bsem1484 (Pcyk-4::CbCYK-4::Kpn-tether::AIR-2(KD)::GFP_TOPO). Overlap PCR was used to merge the zen-4 promoter from bsem1336 and CYK-4::Kpn-tether from bsem1484, and the final product was subcloned into pCR4-TOPOBlunt to create bsem1485 (Pzen-4::CbCYK-4::Kpn tether_TOPOBlunt). The KpnI/SphI fragments from bsem1336 and bsem1485 were ligated together to create bsem1486 (Pzen-4::CYK-4::tether::AIR-2::GFP). The KpnI/SphI fragments from bsem1484 and bsem1485 were ligated together to create bsem1487 (Pzen-4::CYK-4::tether::AIR-2(KD)::GFP). We used both kinase-intact and kinase-dead versions of AIR-2, and observed no rescue with either tether.
Pelt-7::mCherry_pGEM-T (bsem1146), Pdpy-13::mCherry_pGEM-T (bsem1178), and Pelt-7::mCherry::HIS-58/H2B_pGEM-T (bsem1177) were generated by overlap PCR. The 2.9-kb elt-7 promoter was amplified from wild-type genomic DNA and fused with either mCherry::unc-54 3′UTR from pCFJ90 (Addgene plasmid 19327; a gift from Christian Frokjaer-Jansen and Erik Jorgensen, University of Utah; Frokjaer-Jensen et al., 2008) or with mCherry::H2B::unc-54 3′UTR from bsem1145. bsem1145 was generated by releasing mCherry::HIS-58/H2B from pie-1::mCherry::HIS-58/H2B (a gift from Jon Audhya, University of Wisconsin–Madison; McNally et al., 2006) using BamHI (NEB) and subcloning this fragment into the BamHI site of bsem1176 (unc-54_3′UTR_TOPO). The 260–base pair dpy-13 promoter was amplified from wild-type genomic DNA using primers with attB1 and attB2 overhangs. The resulting product was recombined into pDONR221 using BP Clonase II (Invitrogen) to create bsem1184. The dpy-13 promoter was then amplified from bsem1184 and fused with mCherry::H2B::unc-54 3′UTR amplified from bsem1145. The overlap products were then subcloned into pGEM-T (Promega).
Microinjection/transgenic generation
We injected 2 ng of BamHI-linearized ZEN-4::GFP or of the deletion constructs described earlier (as purified overlap PCR products or linearized plasmids digested with either SapI or SphI) into SM1052 (zen-4(px47) dpy-20(e1282)/bli-6(sc16) unc-24(e138)) along with 5–10 ng of linearized bsem1146 (NcoI), bsem1177 (NcoI), and/or bsem1178 (SapI; fluorescent coselectable markers), 55 ng of EcoR1-linearized pRF4 (rol-6(d) coselectable marker), and salmon testes DNA (amount necessary to bring the total DNA in the injection mix to 100 ng; D1626; Sigma-Aldrich) to generate complex array-containing Roller animals.
ZEN-4 tether constructs were generated by in vivo recombination in the nematode gonad. Each half of the tether was generated by PCR and injected in equimolar amounts along with linearized bsem1177, bsem1178, pRF4, and salmon testes DNA.
We injected 2 ng of SphI-linearized bsem1486 into SM1052, along with 10 ng of NcoI-linearized bsem1177, 55 ng of EcoRI-linearized pRF4, and 33 ng of salmon testes DNA (D1626; Sigma-Aldrich) to generate a CYK-4::AIR-2–expressing transgenic line.
We injected 1 ng/μl Pdlg-1–containing purified PCR product (see earlier description), 55 ng/μl linearized pRF4 (rol-6(d) coselection marker), and 44 ng/μl Salmon Testes DNA (D1626; Sigma-Aldrich) into N2 (SM1880) to generate complex array-containing Roller animals.
RNAi
RNAi was performed as described (Timmons et al., 2001), with minor modifications. A single bacterial colony (either mCherry as a negative control or pha-4) was picked to grow in 6 ml of Luria broth plus antibiotic (25 μg/ml kanamycin) for 8 h at 37°C. Bacteria were pelleted and resuspended in an isopropyl-β-d-thiogalactoside (IPTG)/antibiotic mixture (8 mM IPTG, 25 μg/ml kanamycin), and 40 μl was spread onto a 35-mm Nematode Growth Medium (NGM; Stiernagle, 2006) plate. The plates were covered with foil and incubated at room temperature for 48 h before use. N2 gravid adults were bleached to release eggs, which were hatched overnight in M9 without food to synchronize the population. Starved L1s were transferred to OP50-containing NGM plates and grown to the L4 stage at 20°C. L4s were transferred to a conical tube and washed in M9 two or three times to remove all traces of OP50 bacteria. Approximately 80 L4s were dropped onto an mCherry or a pha-4 RNAi plate and incubated at 25°C for 24 h. The now adults were transferred to a new RNAi plate and incubated for an additional 6 h at 25°C. Laid embryos were collected and either incubated overnight at 25°C on NGM plates to score terminal phenotypes or fixed with paraformaldehyde (PFA) immediately for immunostaining.
Live imaging
Embryos were placed on a 4% agar pad (Difco Noble Agar) in 10 mM levamisole. A #1.5 coverslip (Corning) was added, and nail polish was added at the corners to prevent the coverslip from moving. Embryos were scored using Nomarski optics for foregut attachment defects under a Zeiss AxioImager M2 running AxioVision or ZEN2 software. For live imaging of GFP fluorescence, embryos were imaged under either an AxioImager M2 with Apotome or Apotome.2 to remove out-of-focus light or the Zeiss LSM710, LSM780, or LSM880 confocal microscope. For AJM-1::GFP quantification in SU93 and SM1271 animals, z-stacks through the foregut/arcade cells were taken using the same gain and laser power for each embryo. The z-stacks were rotated in Zen Blue software (Zeiss) and culled of any noninformative slices. These modified stacks were imported into Fiji software (Schindelin et al., 2012). The region of interest containing the arcade cells was cropped out of original image and changed to 16 bit, and the stack was z-projected using the Sum Slices setting and then measured to obtain the mean gray value and area. To control for differences in area between samples, the average intensity per pixel was calculated by dividing the mean gray value by the area.
Immunostaining
Embryos were placed on poly-l-lysine (P-8920; Sigma-Aldrich)–coated slides in 50 μl of water and processed using the freeze-crack method (Duerr, 2006; Shakes et al., 2012). After cracking, slides were incubated in ice-cold MeOH for 8 min. The exception was for pha-4(RNAi)–processed embryos, for which the water was removed and replaced by PFA fix (2% PFA diluted from a 16% methanol-free stock [Alfa Aesar] in 1× phosphate-buffered saline [PBS]) and then incubated for 3 min in ice-cold MeOH after cracking. Slides were blocked in TNB buffer (100 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1% bovine serum albumin) with 10% normal goat serum (NGS; 102643-580; VWR). All antibodies were also diluted using TNB plus 10% NGS. The following antibodies were used: mouse anti–ERM-1 (1:10; Developmental Studies Hybridoma Bank [DSHB]; Hadwiger et al., 2010); mouse anti–PAR-3 (1:10; P4A1; DSHB; Nance et al., 2003); rabbit anti-PHA-4 (1:1000; Kaltenbach et al., 2005); mouse anti-PSD95, which detects DLG-1 (MA1-045; 1:250; Pierce; Firestein and Rongo, 2001); rabbit anti–PAR-6 (1:1000, gift from Tony Hyman, Max Planck Institute; Hoege et al., 2010); rabbit anti–AIR-2 (1:200; gift from Jill Schumacher, MD Anderson Cancer Center; Schumacher et al., 1998); rabbit anti–UNC-59 (1:440; gift from Amy Maddox, University of North Carolina at Chapel Hill; Maddox et al., 2005); mouse anti-GFP (1:500; MAB3580; EMD/Millipore); and chicken anti-GFP (1:200; AB16901; EMD/Millipore). As noted, ERM-1 and PAR-3 antibodies were obtained from the DSHB, which was developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology, University of Iowa (Iowa City, IA). Secondary antibodies conjugated to Alexa 488, Alexa 568, or Alexa 647 were obtained from Invitrogen. Images were obtained on a Zeiss LSM780 or 880 confocal microscope. Images were exported from ZEN Blue (Zeiss), and maximum intensity projections were generated in ZEN Black (Zeiss) and rotated, resized, and cropped in Adobe Photoshop.
smFISH procedure and data processing
smFISH was performed mostly as described (Raj and Tyagi, 2010) and is summarized here with some modifications. Custom Stellaris FISH Probes were designed against dlg-1, erm-1, par-6, and gfp by using the Stellaris FISH Probe Designer (Biosearch Technologies, Petaluma, CA; www.biosearchtech.com/stellarisdesigner). Exact probe sequences are given in Supplemental Table S7. dlg-1, erm-1, and par-6 probes were labeled with Quasar 570 dye, pha-4 probes were labeled with CalFluorRed 610 or CalFluorOrange 635, and gfp was labeled with Quasar 670. Probes were resuspended in 400 µl of TE (10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0), aliquoted, and stored at −20°C. Probes in use were stored at 4°C. One to five 60-mm NGM plates with gravid adults and many laid embryos were washed with water to remove adults and larvae into a 15-ml conical tube. To release the laid embryos, additional water was added to the plate and lightly rubbed with a gloved fingertip. After one or two washes in double-distilled H2O (ddH2O), pelleted embryos and worms were transferred to a 1.5-ml centrifuge tube and pelleted. To release the young embryos from the gravid adults, 100–200 μl of bleach solution (240 μl of bleach, 50 μl of 5 M NaOH, and 710 μl of ddH2O) was added to the pellet and incubated for 5 min at room temperature using a thermomixer (Eppendorf) to mix the sample every 30 s. The embryos were quickly washed three times using M9 buffer. The pelleted embryos were then fixed in 100–200 μl of 3.7% formaldehyde solution (100 μl of 37% formaldehyde, 100 μl of 10× PBS, 800 μl of ddH2O) for 15 min at room temperature using a thermomixer to mix the sample every 3 min. The tube was flash-frozen in liquid nitrogen for 1 min to freeze-crack the eggshells and then either stored at −80°C or processed immediately. The frozen sample was thawed at room temperature and then washed twice with 200 μl of 1× PBS. The pelleted embryos were resuspended in 200 μl of 70% EtOH and stored overnight at 4°C. The next day, the EtOH was exchanged for wash buffer (0.5 ml formamide, 0.5 ml 20× saline–sodium citrate [SSC; Invitrogen], 4 ml of ddH2O), and then the sample was pelleted. A 50-μl amount of hybridization buffer (100 μl of deionized formamide [Calbiochem OmniPur/EMD Millipore], 0.1 g of dextran sulfate [Acros], 100 μl of 20× SSC, 800 μl of ddH2O) containing smFISH probes was added to the embryos (0.75 μl for dlg-1 and pha-4 probes, 1 μl for erm-1 and par-6 probes, and 2 μl for gfp probes) and incubated for 4 h at 37°C in a thermomixer, with mixing every 15 min. The embryos were rinsed once quickly using wash buffer and then washed for 1 h at 37°C in a thermomixer (mixing every 5 min). Pelleted embryos were resuspended in 30–50 μl of ddH2O and then added to polylysine-coated slides and allowed to settle. The ddH2O was removed, and 7 μl of SloFade with 4′,6-diamidino-2-phenylindole (DAPI) was added. A #1.5 coverslip was added, excess mounting material was wicked away with a Kimwipe, and it was sealed with clear nail polish. Slides were imaged immediately or stored at −20°C until ready to image. Images were obtained on a Zeiss LSM880 confocal microscope with a Plan-Apochromat 63×/1.4 oil objective. For imaging dlg-1 and gfp transcripts, 568- and 633-nm laser lines were used. To spectrally distinguish Quasar 570 and CalFluorRed610 or CalFluorOrange635 (e.g., imaging dlg-1 and pha-4 in same embryo), we excited Quasar570 with a 514-nm laser and adjusted the GAASP detector range on the LSM880 to acquire emission wavelengths between 561 and 596 nm; we excited CalFluorRed610 or CalFluorOrange635 with a 594-nm laser and acquired emission wavelengths between 623 and 694 nm. GFP protein expression survived the fixation method and could be visualized with a 488-nm laser. Selected z-stacks that encompassed the digestive tract but excluded the epidermis were exported from ZEN Blue (Zeiss) and imported into Imaris 3D software (www.bitplane.com) for converting the raw smFISH signal into painted spheres. For quantitation of transcripts in the arcade cells, zen-4 dpy-20/mIs11 (SM2300) embryos were processed as described and hybridized with pha-4 probes plus dlg-1, par-6, or erm-1 probes. The zen-4/+ versus zen-4 mutant embryos were determined by the presence or absence, respectively, of GFP expressed from the balancing transgene. Embryos between comma and 1.25-fold (stages before or after arcade cell polarization) were imaged using a Zeiss LSM880 confocal. Z-stacks encompassing the arcade cells (identified as the cells anterior to the foregut that expressed pha-4 RNA) were selected and imported into Imaris for quantitation. Only the region corresponding to the arcade cells was quantified. To determine statistical significance, the Wilcoxon rank-sum test was used in GraphPad Prism. In all scatter plots, the mean is shown by the long horizontal line and the 95% confidence intervals by the shorter horizontal lines. For dlg-1 and associated pha-4 quantitation, p = 0.9312 and 0.1636, respectively. For par-6 and associated pha-4 quantitation, p = 0.0862 and 0.1171, respectively. For erm-1 and associated pha-4 quantitation, p = 0.9551 and 0.1453, respectively.
PHA-4 and FoxA ChIP-seq analysis
For the C. elegans analysis, bed files from the early and late embryo PHA-4 ChIP-seq data sets (as well as the MACS2 peak analysis bed file; Zhong et al., 2010) and the Ahringer and Meyer TSS data sets (Chen et al., 2013; Kruesi et al., 2013) were downloaded and visualized in the Broad Institute Integrative Genomics Viewer (Robinson et al., 2011; Thorvaldsdottir et al., 2013). Genes known to be expressed in epithelial tissue (those listed in Table 1 and Supplemental Table S1) were queried, and a gene was annotated as PHA-4 bound if a MACS2-called PHA-4 peak was found upstream, at the TSS, or within the first intron.
For the mammalian analysis, FoxA1/FoxA2 ChIP-seq data were downloaded as peak calls for the following data sets: FoxA2 ChIP-seq from mouse liver (Soccio et al., 2011); FoxA1 ChIP-seq from HEPG2 cells (Motallebipour et al., 2009); FoxA2 ChIP-seq from Caco2 cells (Gosalia et al., 2015); and FoxA1 and FoxA2 from human embryonic stem cells differentiated into gut tube or foregut cells (Wang et al., 2015). For the Caco2 data set, two replicates were downloaded, and the intersection was used for subsequent analysis. For FoxA1 ChIP-seq from MCF7 cells (Hurtado et al., 2011), two replicates in FASTQ format were obtained and then processed to call peaks. Quality trim of reads was performed using Trimmomatic (Bolger et al., 2014), mapping of treatment replicates and control input to genome was performed using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; Langmead and Salzberg, 2012), and peaks were called using MACS2 (https://pypi.python.org/pypi/MACS2; Zhang et al., 2008). The intersection of the replicate peaks was generated and used for subsequent analysis.
Where needed, the input files were lifted to either mouse mm10 (GRCm38) or human hg38 (GRCh38) genomes using the UCSC LiftOver program (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Mouse and human gene annotation, as well as mouse and human orthologues of C. elegans genes, were downloaded from Ensembl (release 80). FoxA peaks that were up to 1000 base pairs upstream, up to 1000 base pairs downstream, or within the gene body of mouse and human genes were identified, and the final output was made up of homologues of the 50 C. elegans PHA-4–bound, epithelial-specific genes (Table 1) that had FoxA binding as per these parameters. A manual inspection was performed to determine the percentage of genes that had FoxA binding upstream or within the first or second intron. An affirmative call did not exclude binding in other areas beyond upstream or introns 1 and 2.
Supplementary Material
Acknowledgments
We thank J. Nance for par-6(MZ) reagents; the Developmental Studies Hybridoma Bank, T. Hyman, J. Schumacher, and A. Maddox for antibodies; D. Zhang and M. Glotzer for sharing ect-2(xs110) before publication; M. Labouesse for DLG-1::GFP plasmids and worm strains; C. Rongo, E. Jorgensen, C. Frokjaer-Jansen, and J. Audhya for plasmids; D. Richardson at the Harvard Center for Biological Imaging for help optimizing smFISH imaging; J. Von Stetina for help with GraphPad Prism; J. Lisack and L. Rosen for technical help; and R. Losick and A. Schier for comments on the manuscript. Some strains were obtained from the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Grant P40 OD010440. Funding was supplied by Harvard University, the University of Utah, the MacArthur Foundation, and National Institutes of Health Grants 5F32 GM084650 (S.E.V.S.), 1R21 HD066263 (S.E.M.), and 5R37 GM056264 (S.E.M.).
Abbreviations used:
- ChIP-seq
chromatin immunoprecipitation sequencing
- CYK
cytokinesis defective
- DLG
Discs Large
- EMT
epithelial-to-mesenchymal transition
- ERM
ezrin/radixin/moesin homologue
- FoxA
forkhead box transcription factor subgroup A
- MET
mesenchymal-to-epithelial transition
- PAR
partitioning defective
- PHA
defective pharynx development
- smFISH
single-molecule fluorescence in situ hybridization
- TSS
transcription start site
- ZEN
zygotic enclosure defective.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0644) on May 24, 2017.
REFERENCES
- Achilleos A, Wehman AM, Nance J. PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development. 2010;137:1833–1842. doi: 10.1242/dev.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Hernandez X, Nichols GM, Glass J. Caco-2 cell line: a system for studying intestinal iron transport across epithelial cell monolayers. Biochim Biophys Acta. 1991;1070:205–208. doi: 10.1016/0005-2736(91)90165-5. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bersaas A, Arnoldussen YJ, Sjøberg M, Haugen A, Mollerup S. Epithelial-mesenchymal transition and FOXA genes during tobacco smoke carcinogen induced transformation of human bronchial epithelial cells. Toxicol In Vitro. 2016;35:55–65. doi: 10.1016/j.tiv.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Besnard V, Wert SE, Hull WM, Whitsett JA. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr Patterns. 2004;5:193–208. doi: 10.1016/j.modgep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bi E, Park H-O, Abe M, Qadota H, Hirata A, Ohya Y, Adames N, Blundell K, Ashby MN, Boone C, et al. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J Biol Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Bossinger O, Fukushige T, Claeys M, Borgonie G, McGhee JD. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev Biol. 2004;268:448–456. doi: 10.1016/j.ydbio.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Bossinger O, Klebes A, Segbert C, Theres C, Knust E. Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev Biol. 2001;230:29–42. doi: 10.1006/dbio.2000.0113. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breznau EB, Semack AC, Higashi T, Miller AL. MgcRacGAP restricts active RhoA at the cytokinetic furrow and both RhoA and Rac1 at cell-cell junctions in epithelial cells. Mol Biol Cell. 2015;26:2439–2455. doi: 10.1091/mbc.E14-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodu V, Casanova J. The RhoGAP crossveinless-c links trachealess and EGFR signaling to cell shape remodeling in Drosophila tracheal invagination. Genes Dev. 2006;20:1817–1828. doi: 10.1101/gad.375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 2008;322:1543–1546. doi: 10.1126/science.1163086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Castanon I, Von Stetina S, Kass J, Baylies MK. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128:3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- Caviglia S, Luschnig S. Tube fusion: making connections in branched tubular networks. Semin Cell Dev Biol. 2014;31:82–90. doi: 10.1016/j.semcdb.2014.03.018. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- Chen M-C, Zhou Y, Detrich HW, 3rd Zebrafish mitotic kinesin-like protein 1 (Mklp1) functions in embryonic cytokinesis. Physiol Genomics. 2002;8:51–66. doi: 10.1152/physiolgenomics.00042.2001. [DOI] [PubMed] [Google Scholar]
- Chen RA-J, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res. 2013;23:1339–1347. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Chisholm AD, Hardin J. Epidermal morphogenesis. WormBook. 2005;2005(Dec 1):1–22. doi: 10.1895/wormbook.1.35.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes AN, Davies JA, Little MH. Cell-cell interactions driving kidney morphogenesis. Curr Top Dev Biol. 2015;112:467–508. doi: 10.1016/bs.ctdb.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C. A Putative catenin–cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deavours BE, Walker RA. Nuclear localization of C-terminal domains of the kinesin-like protein MKLP-1. Biochem Biophys Res Commun. 1999;260:605–608. doi: 10.1006/bbrc.1999.0952. [DOI] [PubMed] [Google Scholar]
- del Castillo U, Lu W, Winding M, Lakonishok M, Gelfand VI. Pavarotti/MKLP1 regulates microtubule sliding and neurite outgrowth in Drosophila neurons. Curr Biol. 2015;25:200–205. doi: 10.1016/j.cub.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne LK, Wang X-J, Prekeris R. Midbody: from cellular junk to regulator of cell polarity and cell fate. Curr Opin Cell Biol. 2015;35:51–58. doi: 10.1016/j.ceb.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr JS. Immunohistochemistry. WormBook. 2006;2006(Jun 19):1–61. doi: 10.1895/wormbook.1.105.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- Firestein BL, Rongo C. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Mol Biol Cell. 2001;12:3465–3475. doi: 10.1091/mbc.12.11.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The FoxA family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Joberty G, Macara IG. Assembly of epithelial tight junctions is negatively regulated by Par6. Curr Biol. 2002;12:221–225. doi: 10.1016/s0960-9822(01)00663-7. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalia N, Yang R, Kerschner JL, Harris A. FOXA2 regulates a network of genes involved in critical functions of human intestinal epithelial cells. Physiol Genomics. 2015;47:290–297. doi: 10.1152/physiolgenomics.00024.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot L, Guerrera D, Spadaro D, Tapia R, Jond L, Citi S. MgcRacGAP interacts with cingulin and paracingulin to regulate Rac1 activation and development of the tight junction barrier during epithelial junction assembly. Mol Biol Cell. 2014;25:1995–2005. doi: 10.1091/mbc.E13-11-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger G, Dour S, Arur S, Fox P, Nonet ML. A monoclonal antibody toolkit for C. elegans. PLoS One. 2010;5:e10161. doi: 10.1371/journal.pone.0010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra KM, Chen DY-S, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- Hardin J, King R, Thomas-Virnig C, Raich WB. Zygotic loss of ZEN-4/MKLP1 results in disruption of epidermal morphogenesis in the C. elegans embryo. Dev Dyn. 2008;237:830–836. doi: 10.1002/dvdy.21455. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid PJ, Raich WB, Smith R, Mohler WA, Simokat K, Gendreau SB, Rothman JH, Hardin J. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev Biol. 2001;236:165–180. doi: 10.1006/dbio.2001.0315. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hoege C, Constantinescu AT, Schwager A, Goehring NW, Kumar P, Hyman AA. LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr Biol. 2010;20:1–8. doi: 10.1016/j.cub.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Kolodziej PA. Organogenesis: molecular mechanisms of tubulogenesis. Nat Rev Genet. 2002;3:513–523. doi: 10.1038/nrg840. [DOI] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H-T, Chen H-M, Yang Z, Wang J, Lee NK, Burger A, Zaret K, Liu T, Levine E, Mango SE. Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science. 2015;348:1372–1376. doi: 10.1126/science.aab1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Ji N, van Oudenaarden A. Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. WormBook. 2012;2012(Dec 13):1–16. doi: 10.1895/wormbook.1.153.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungkamp A-C, Stoeckius M, Mecenas D, Grün D, Mastrobuoni G, Kempa S, Rajewsky N. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell. 2011;44:828–840. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach LS, Updike DL, Mango SE. Contribution of the amino and carboxyl termini for PHA-4/FoxA function in Caenorhabditis elegans. Dev Dyn. 2005;234:346–354. doi: 10.1002/dvdy.20550. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kelley M, Yochem J, Krieg M, Calixto A, Heiman MG, Kuzmanov A, Meli V, Chalfie M, Goodman MB, Shaham S, et al. FBN-1, a fibrillin-related protein, is required for resistance of the epidermis to mechanical deformation during C. elegans embryogenesis. Elife. 2015;4:1–30. doi: 10.7554/eLife.06565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmanov A, Yochem J, Fay DS. Analysis of PHA-1 reveals a limited role in pharyngeal development and novel functions in other tissues. Genetics. 2014;198:259–268. doi: 10.1534/genetics.114.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-H, Schedl T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 2004;18:1047–1059. doi: 10.1101/gad.1188404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi M-H, Médina E, Arsanto J-P, Le Bivic A. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15:1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Loria A, Longhini KM, Glotzer M. The RhoGAP domain of CYK-4 has an essential role in RhoA activation. Curr Biol. 2012;22:213–219. doi: 10.1016/j.cub.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox AS, Habermann B, Desai A, Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- Mango SE. The molecular basis of organ formation: insights from the C. elegans foregut. Annu Rev Cell Dev Biol. 2009;25:597–628. doi: 10.1146/annurev.cellbio.24.110707.175411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Matuliene J, Kuriyama R. Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol Biol Cell. 2002;13:1832–1845. doi: 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley D, Wienhold M, Sun X. The pulmonary mesenchyme directs lung development. Curr Opin Genet Dev. 2015;32:98–105. doi: 10.1016/j.gde.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown C, Praitis V, Austin J. sma-1 encodes a betaH-spectrin homolog required for Caenorhabditis elegans morphogenesis. Development. 1998;125:2087–2098. doi: 10.1242/dev.125.11.2087. [DOI] [PubMed] [Google Scholar]
- McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114:2265–2277. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier V, Bourrié M, Berger Y, Fabre G. The human intestinal epithelial cell line Caco-2; pharmacological and pharmacokinetic applications. Cell Biol Toxicol. 1995;11:187–194. doi: 10.1007/BF00756522. [DOI] [PubMed] [Google Scholar]
- Minestrini G, Harley AS, Glover DM. Localization of Pavarotti-KLP in living Drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol Biol Cell. 2003;14:4028–4038. doi: 10.1091/mbc.E03-04-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minestrini G, Mathe E, Glover DM. Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: Effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J Cell Sci. 2002;115:725–736. doi: 10.1242/jcs.115.4.725. [DOI] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Motallebipour M, Ameur A, Reddy Bysani MS, Patra K, Wallerman O, Mangion J, Barker MA, McKernan KJ, Komorowski J, Wadelius C. Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq. Genome Biol. 2009;10:R129. doi: 10.1186/gb-2009-10-11-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Munro EM, Priess JR. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–5350. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Dickinson DJ, Weis WI. Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog Mol Biol Transl Sci. 2013;116:3–23. doi: 10.1016/B978-0-12-394311-8.00001-7. [DOI] [PubMed] [Google Scholar]
- Neuhold LA, Wold B. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Sawa H, Okano H, White JG. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci. 2000;113:3825–3837. doi: 10.1242/jcs.113.21.3825. [DOI] [PubMed] [Google Scholar]
- Pásti G, Labouesse M. Epithelial junctions, cytoskeleton, and polarity. WormBook. 2014;2014(Nov 4):1–35. doi: 10.1895/wormbook.1.56.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol Biol Cell. 2007;18:4992–5003. doi: 10.1091/mbc.E07-05-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz B, White JG. Cell fusions in the developing epithelial of C. elegans. Dev Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Pollarolo G, Schulz JG, Munck S, Dotti CG. Cytokinesis remnants define first neuronal asymmetry in vivo. Nat Neurosci. 2011;14:1525–1533. doi: 10.1038/nn.2976. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Mango SE. Early morphogenesis of the Caenorhabditis elegans pharynx. Dev Biol. 2001;233:482–494. doi: 10.1006/dbio.2001.0235. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Saam J, Mango SE. ZEN-4/MKLP1 is required to polarize the foregut epithelium. Curr Biol. 2004;14:932–941. doi: 10.1016/j.cub.2004.05.052. [DOI] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- Raich WB, Agbunag C, Hardin J. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr Biol. 1999;9:1139–1146. doi: 10.1016/S0960-9822(00)80015-9. [DOI] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Tyagi S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 2010;472:365–386. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- Rasmussen JP, Reddy SS, Priess JR. Laminin is required to orient epithelial polarity in the C. elegans pharynx. Development. 2012;139:2050–2060. doi: 10.1242/dev.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A, Gomez GA, Priya R, Verma S, Kovacs EM, Jiang K, Brown NH, Akhmanova A, Stehbens SJ, Yap AS. Centralspindlin and alpha-catenin regulate Rho signalling at the epithelial zonula adherens. Nat Cell Biol. 2012;14:818–828. doi: 10.1038/ncb2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E, Bishop JD, Waddle JA, Schumacher JM, Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157:219–229. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Fan S, Liu C-JJ, Margolis B. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci. 2003;116:2895–2906. doi: 10.1242/jcs.00500. [DOI] [PubMed] [Google Scholar]
- Roignot J, Peng X, Mostov K. Polarity in mammalian epithelial morphogenesis. Cold Spring Harb Perspect Biol. 2013;5:a013789. doi: 10.1101/cshperspect.a013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TL, Hashimoto S, Gu SG, Morton JJ, Stadler M, Blumenthal T, Fire A, Morishita S. The transcription start site landscape of C. elegans. Genome Res. 2013;23:1348–1361. doi: 10.1101/gr.151571.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher JM, Golden A, Donovan PJ. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C, Johnson K, Theres C, van Furden D, Bossinger O. Molecular and functional analysis of apical junction formation in the gut epithelium of Caenorhabditis elegans. Dev Biol. 2004;266:17–26. doi: 10.1016/j.ydbio.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Severson AF, Hamill DR, Carter JC, Schumacher J, Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Miller DM, Nonet ML. Immunofluorescence microscopy. Methods Cell Biol. 2012;107:35–66. doi: 10.1016/B978-0-12-394620-1.00002-3. [DOI] [PubMed] [Google Scholar]
- Shaye DD, Casanova J, Llimargas M. Modulation of intracellular trafficking regulates cell intercalation in the Drosophila trachea. Nat Cell Biol. 2008;10:964–970. doi: 10.1038/ncb1756. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PVE, Kamath RS, Fraser AG, Ahringer J, Plasterk RHA. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Pohl C. Coupling of rotational cortical flow, asymmetric midbody positioning, and spindle rotation mediates dorsoventral axis formation in C. elegans. Dev Cell. 2014;28:253–267. doi: 10.1016/j.devcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol. 2009;1:a003384. doi: 10.1101/cshperspect.a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol Endocrinol. 2011;25:694–706. doi: 10.1210/me.2010-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume A-M, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006;2006(Feb 11):1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tang Y, Shu G, Yuan X, Jing N, Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21:316–326. doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K, Furihata M, Saibara T. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia. 2014;16:1082–1093. doi: 10.1016/j.neo.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Fasulo B, Warecki B, Sullivan W, Verhey KJ. Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor. Nat Commun. 2016;7:11182. doi: 10.1038/ncomms11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–1268. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- Updike DL, Mango SE. Genetic suppressors of Caenorhabditis elegans pha-4/FoxA identify the predicted AAA helicase ruvb-1/RuvB. Genetics. 2007;177:819–833. doi: 10.1534/genetics.107.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe KJ, White JG. Cortical centralspindlin and G alpha have parallel roles in furrow initiation in early C. elegans embryos. J Cell Sci. 2007;120:1772–1778. doi: 10.1242/jcs.03447. [DOI] [PubMed] [Google Scholar]
- Von Stetina SE, Mango SE. PAR-6, but not E-cadherin and β-integrin, is necessary for epithelial polarization in C. elegans. Dev Biol. 2015;403:5–14. doi: 10.1016/j.ydbio.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora M, Shah M, Ostafi S, Onken B, Xue J, Ni JZ, Gu S, Driscoll M, Kenyon C, Tissenbaum H, et al. Deletion of microRNA-80 activates dietary restriction to extend C. elegans Healthspan and Lifespan. PLoS Genet. 2013;9:e1003737. doi: 10.1371/journal.pgen.1003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong-Brender TTK, Yang X, Labouesse M. C. elegans Embryonic Morphogenesis. Curr Topics Dev Biol. 2016;116:597–616. doi: 10.1016/bs.ctdb.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Waaijers S, Ramalho JJ, Koorman T, Kruse E, Boxem M. The C. elegans Crumbs family contains a CRB3 homolog and is not essential for viability. Biol Open. 2015;4:276–284. doi: 10.1242/bio.201410744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhu C-P, Hu P-F, Qian H, Ning B-F, Zhang Q, Chen F, Liu J, Shi B, Zhang X, et al. FOXA2 suppresses the metastasis of hepatocellular carcinoma partially through matrix metalloproteinase-9 inhibition. Carcinogenesis. 2014;35:2576–2583. doi: 10.1093/carcin/bgu180. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskelet. 2012;69:882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EA, Raghuraman H, Perozo E, Glotzer M. Binding of the CYK-4 subunit of the centralspindlin complex induces a large scale conformational change in the kinesin subunit. J Biol Chem. 2013;288:19785–19795. doi: 10.1074/jbc.M113.463695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124:2889–2901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Grawe F, Knust E. CRUMBS is involved in the control of apical protein targeting during Drosophila epithelial development. Mech Dev. 1993;44:175–187. doi: 10.1016/0925-4773(93)90066-7. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yang X, Pursell B, Lu S, Chang T-K, Mercurio AM. Regulation of beta 4-integrin expression by epigenetic modifications in the mammary gland and during the epithelial-to-mesenchymal transition. J Cell Sci. 2009;122:2473–2480. doi: 10.1242/jcs.049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Glotzer M. The RhoGAP activity of CYK-4/MgcRacGAP functions non-canonically by promoting RhoA activation during cytokinesis. Elife. 2015;4:e08898. doi: 10.7554/eLife.08898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang C, Gao W, Chen T, Qian T, Hu J, Tan Y. FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer. Cancer Lett. 2015;361:240–250. doi: 10.1016/j.canlet.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HYK, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravlev Y, Hirsch SM, Jordan SN, Dumont J, Shirasu-Hiza M, Canman JC. CYK-4 regulates Rac, but not Rho, during cytokinesis. Mol Biol Cell. 2017;28:1258–1270. doi: 10.1091/mbc.E17-01-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.