FIGURE 7:
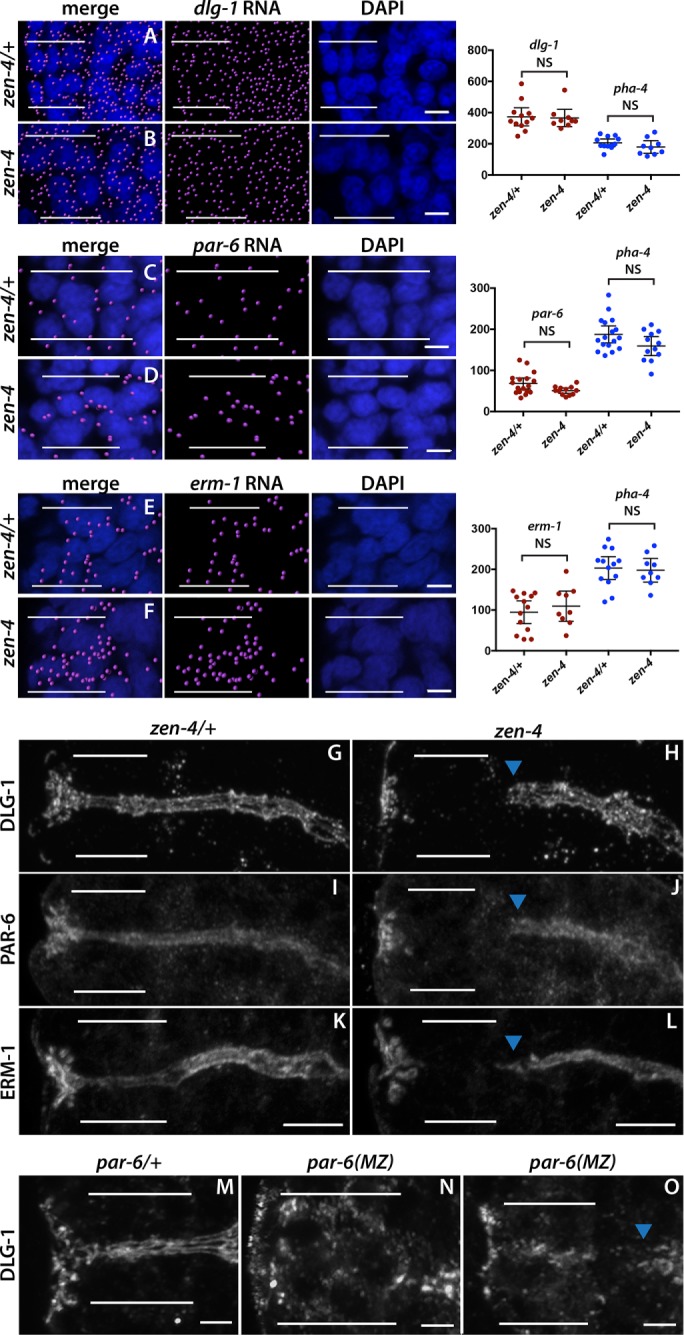
zen-4 and par-6 are required for arcade cell polarization. (A–F) mRNA detected by smFISH was imported into Imaris 3D software and is pseudocolored magenta (Materials and Methods); DAPI is blue. Epithelial factor mRNAs (dlg-1, par-6, erm-1) are present in both zen-4/+ (A, C, E) and zen-4 mutant (B, D, F) arcade cells, which are highlighted by white bars. As shown on the graphs on the right, no significant difference in the amount of RNA between zen-4/+ and zen-4 mutant arcade cells was detected (p > 0.05 for all comparisons between control and mutant; see Materials and Methods for actual values). pha-4 RNA was used as a negative control and to help localize the arcade cells for quantification and was also unchanged between zen-4/+ and zen-4 arcade cells (p > 0.05, see Materials and Methods for actual values). (G–L) PAR-6 (G), DLG-1 (I), and ERM-1 (K) are expressed and localized to the apical/junctional domain in zen-4/+ arcade cells (white bars). These proteins are not detected in zen-4 mutant arcade cells (H, J, L) but are expressed and localized in the neighboring foregut (arrowhead). (M–O) DLG-1 is expressed and localized in the arcade cells (white bars) of par-6/+ (M) embryos. Forty percent (14 of 35) of par-6 mutant embryos contain DLG-1, but it is mislocalized (N). In the other 60% (21of 35), DLG-1 is properly localized but fails to form continuous junctions, similar to the adjacent foregut (blue arrowhead; O). All images are maximum intensity projections. Anterior is to the left. Scale bars, 3 μm (A–F), 5 μm (G–L), 2 μm (M–O).
