Figure S3.
Vector Properties Determine Homologous Integration Rates, Related to Figure 2
Vector-specific integration efficiencies were calculated for the set of 915 dispensable genes by normalizing the relative abundance of a mutant during the infection to the relative abundance of the vector measured from the electroporation cuvette, and by using the four normally growing controls to normalize between experiments.
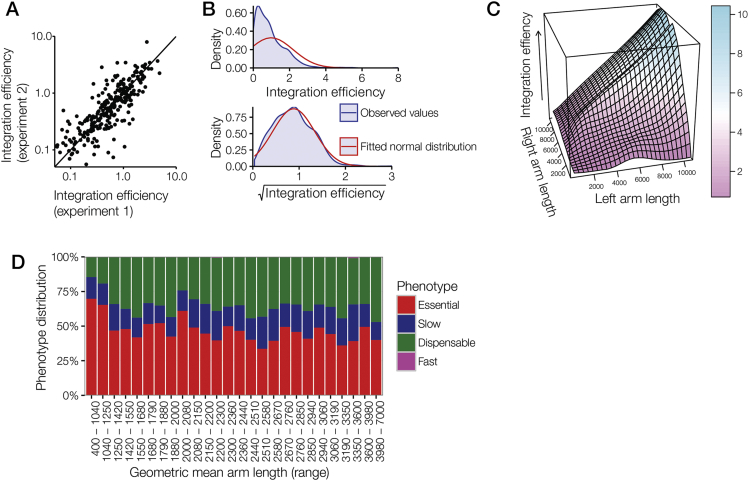
(A) Vector integration efficiencies were highly reproducible between independent experiments (log-log R2: 0.76).
(B) Relative abundances of dispensable mutants became normally distributed after applying a square root function, suggesting targeting efficiency might be the result of two independent variables interacting in a multiplicative fashion.
(C) Modeling the effect on targeting efficiency of homology arm lengths, which in the PlasmoGEM resource varies from 400 bp to 14.8 kb. Initial analyses revealed the length of each homology arm to be independently linked with integration efficiency. This effect plateaus at around 5 kb due to the confounding fact that the lengths of the two homology arms are inversely correlated, since they trade off against each other for space on the vector. The graph shows a three dimensional model fitted to the data, and illustrates increasing targeting efficiency of vectors with arm length up to at least 10 kb. The product of homology arms lengths explained around 60% of the overall variation in targeting efficiency (log-log R2: 0.42). The remaining non-stochastic variation may be due to DNA structure and chromatin state, but combining a number of data sources with machine learning approaches failed to model these factors to improve predictive accuracy.
(D) Assessment of calculated phenotypes across a range of geometric-mean homology arm lengths (groups are, as far as possible, of equal sizes). There is an even phenotype distribution across the space of homology arms, with the possible exception of a technical bias toward essential calls for vectors with a geometric mean homology arm length less than 1.25 kb. As a result, this set of vectors was discarded when calculating overall genome essentiality.