Figure 1.
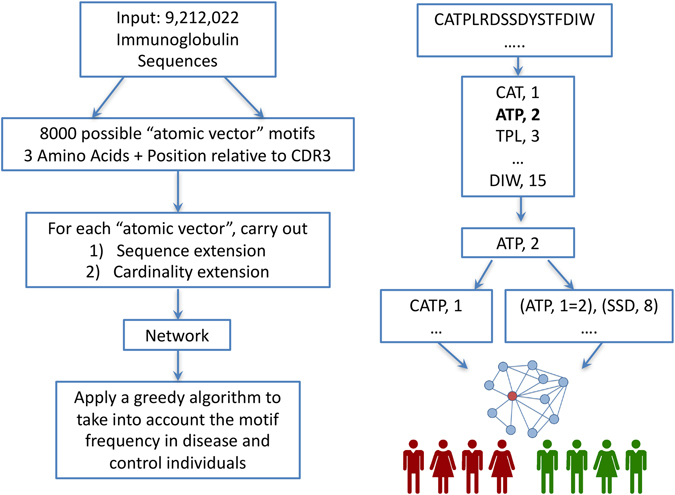
Methodology Overview and Example. (A) We start with a set of over 9 million immunoglobulin sequences in amino acid space. The sequences are then broken down to 8,000 (20^3) atomic vectors of size three. For each of the possible vectors we carry out up to three sequence and cardinality extension steps and apply a greedy algorithm to examine the presence of the motif in the MS and healthy individuals. The traversal is stopped if it is not promising based on a threshold. (B) Schematic illustration of constructing atomic vectors from a single read sequence. See Haystack Heuristic Overview in the Methods section for a detailed explanation.
