Figure 3. RPA stimulates XPF‐ERCC1 activity by binding to the 5′ arms of a DNA fork substrate.
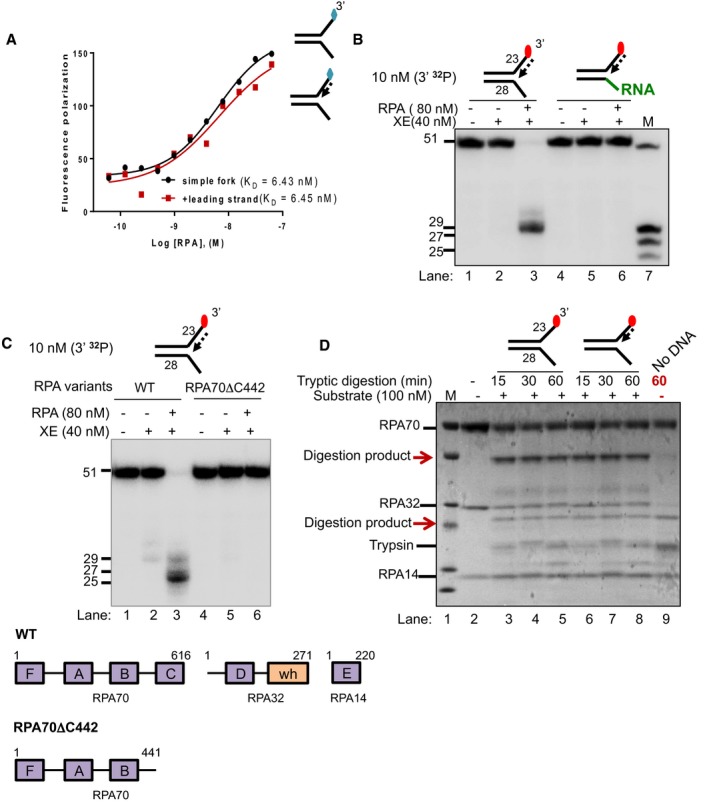
- Fluorescence anisotropy assay to determine the binding constants of RPA for either “simple fork” or “+leading‐strand” substrates. The blue diamonds denote the fluorophore‐labelled nucleotides.
- Nuclease activity of XE on “+leading strand” or DNA:RNA hybrid (5′ ssRNA on the bottom strand) substrates in the presence or absence of 80 nM RPA. RPA cannot stimulate XE to overcome the inhibition of a model nascent leading strand when the 5′‐ssDNA overhang is replaced with 5′ ssRNA. Green line denotes RNA.
- (Top panel) Nuclease activity of XE on “+leading‐strand” substrate in the presence or absence of either the WT RPA or the truncated RPA (RPA70C442). (Bottom panel) A schematic representation of the structural domains of WT RPA and RPA70C442. Purple boxes represent the DNA‐binding domains (DBD) designated as A–F. The orange box represents the winged helix domain. RPA70, RPA32 and RPA14 denote the three subunits of RPA.
- Limited proteolysis assay to determine structural changes in RPA in the presence or absence of the indicated substrates. 800 nM RPA was incubated with 100 nM unlabelled DNA substrates (simple fork; +leading strand; or no DNA) prior to digestion with 500 nM trypsin in a time course. Reaction samples were separated in Bis‐Tris SDS–PAGE (4–12%) and stained with InstantBlue. Red arrows indicated tryptic digestion pattern of RPA.
