Figure EV4. RPA stimulation of XPF‐ERCC1 activity on “+leading‐strand” structure is not attributed to the displacement of the model nascent leading strand or the unwinding of the fork substrates by RPA.
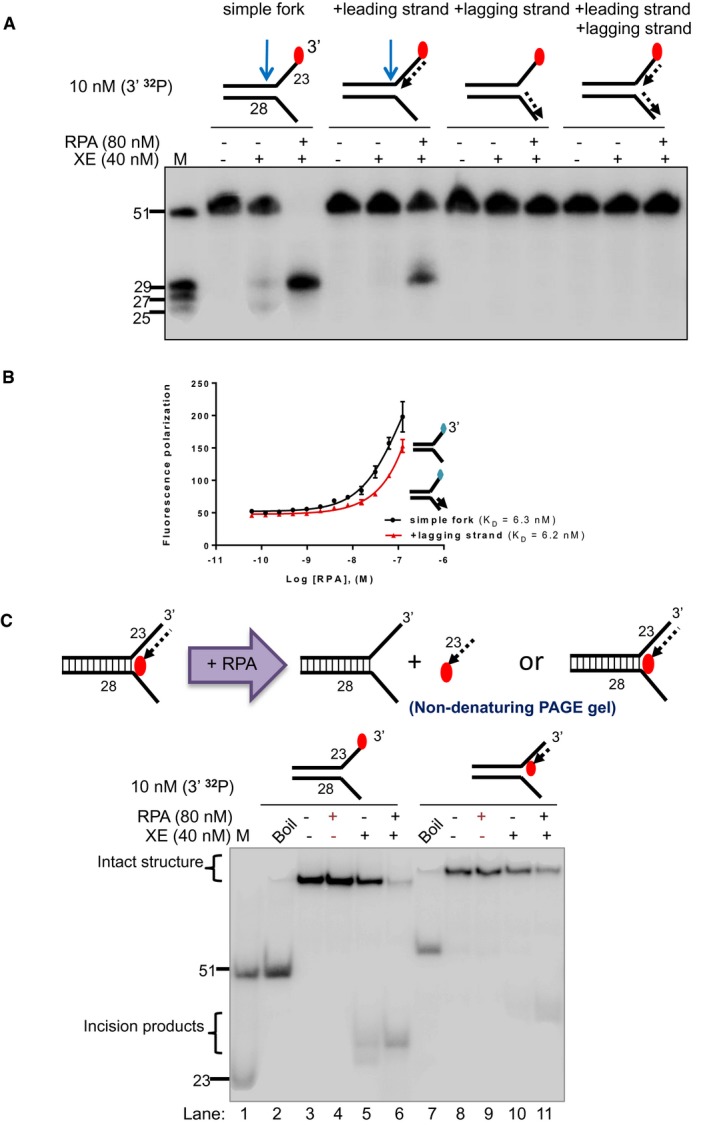
- Nuclease activity of XE on the indicated fork substrates in the presence or absence of RPA. RPA specifically stimulates XE activity on a “simple fork” and “+leading‐strand” substrates.
- Fluorescence anisotropy assay to determine the binding constants of RPA for either “simple fork” or “+lagging‐strand” substrates. The blue diamonds denote the fluorophore‐labelled nucleotides. Error bars represent SD, n = 3.
- (Top panel) Outline of potential consequences of incubating “+leading‐strand” substrate radiolabelled on the model nascent leading strand with RPA, and the potential products that might be revealed by analysis on a non‐denaturing PAGE gel. (Bottom panel) Nuclease activity performed as in panel a. Reaction products were separated on a 10% non‐denaturing PAGE gel. The DNA substrates remain intact in the RPA alone reactions (lanes 4 and 9), indicating that RPA does not displace the model nascent leading strand or unwind the fork substrates, at the concentrations employed.
Source data are available online for this figure.
