Figure 5. The 5′–3′ exonuclease SNM1A can load from an incision induced by XPF‐ERCC1 to digest past a crosslink.
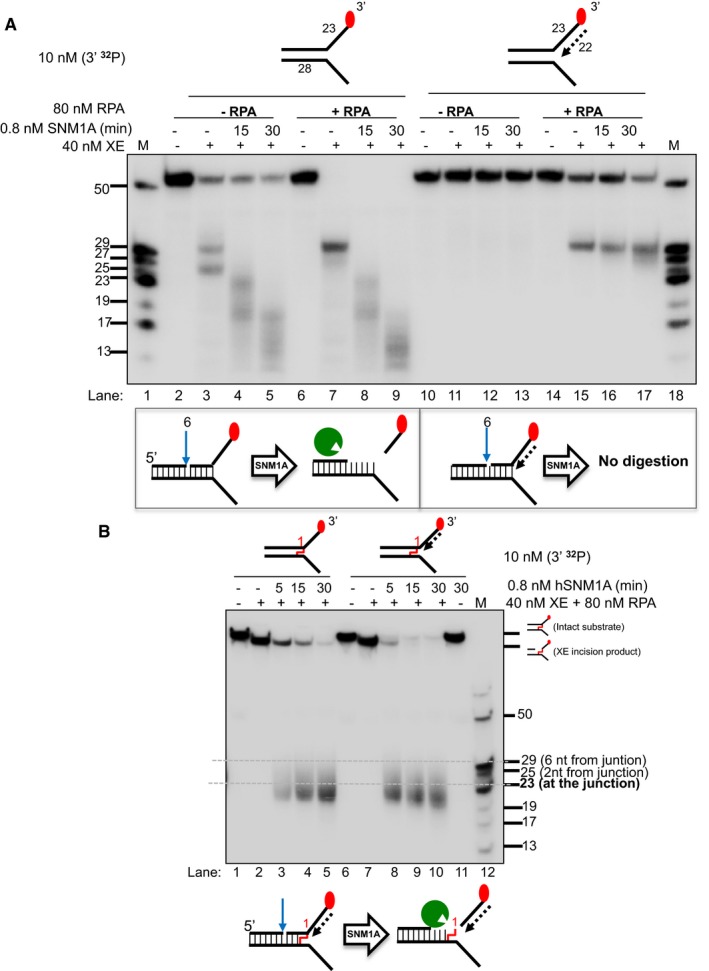
- (Top panel) Nuclease activity of XE on “simple fork” and +leading‐strand substrates in the presence or absence of RPA and further incubated with 0.8 nM SNM1A in a time course. (Bottom panel) Schematic representation of the nuclease assay reaction products. The blue arrow denotes incision by XE and green Pacman represents digestion by SNM1A. RPA does not affect/alter SNM1A exonuclease activity as seen in the similar stepwise digestion products of SNM1A in the presence or absence of RPA (lanes 2–9). However, the presence of a model leading strand prevents SNM1A from loading onto XE‐RPA‐induced incisions to digest the DNA substrate (lanes 10–17).
- (Top panel) Nuclease activity of XE‐RPA on crosslinked DNA substrates (simple fork; +leading strand) and further incubation with SNM1A in a time course. SNM1A digestion inhibition by a model nascent leading strand (as in A) is overcome when an ICL is located at the fork junction.
