Fig. 10.
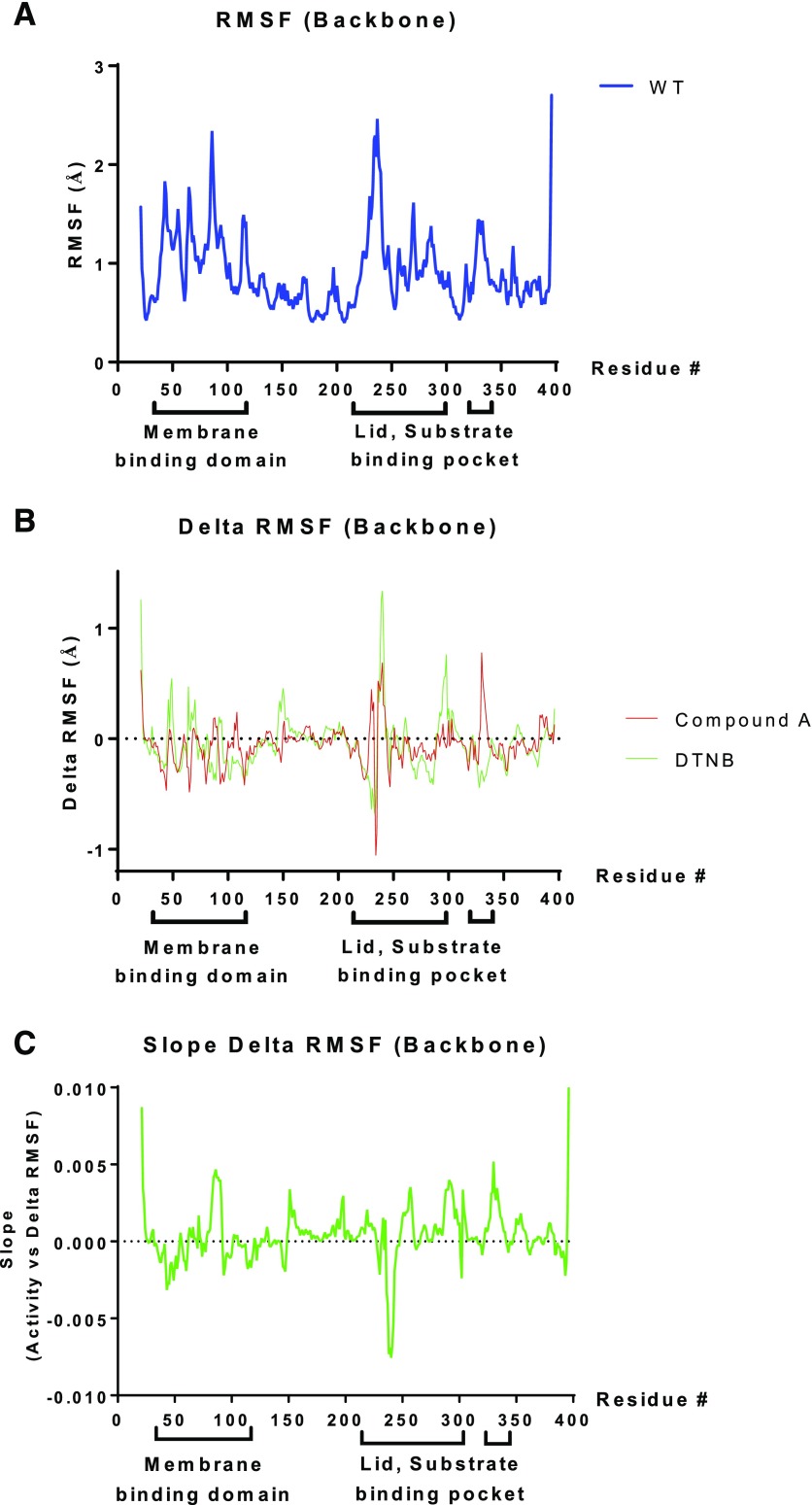
Molecular dynamics simulations of LCAT reveal backbone motion. (A) RMSF (in Ångstrom units) for WT-LCAT compared with the starting structure at each position in the LCAT sequence. (B) The ΔRMSF of WT-LCAT modified either with DTNB (green) or compound A (red) was calculated as the difference from the RMSF of WT-LCAT. (C) Correlation between ΔRMSF with LCAT activity, with a positive slope corresponding to protein regions in which increased backbone motion is associated with increased LCAT activity. Amino acid residues in the membrane binding domain (36–101) and the cap domain containing the lid (residues 226–246), as well as residues that help shape the substrate binding pocket (Glukhova et al., 2015; Piper et al., 2015), are shown below the x-axis. RMSF, root-mean-square fluctuation.