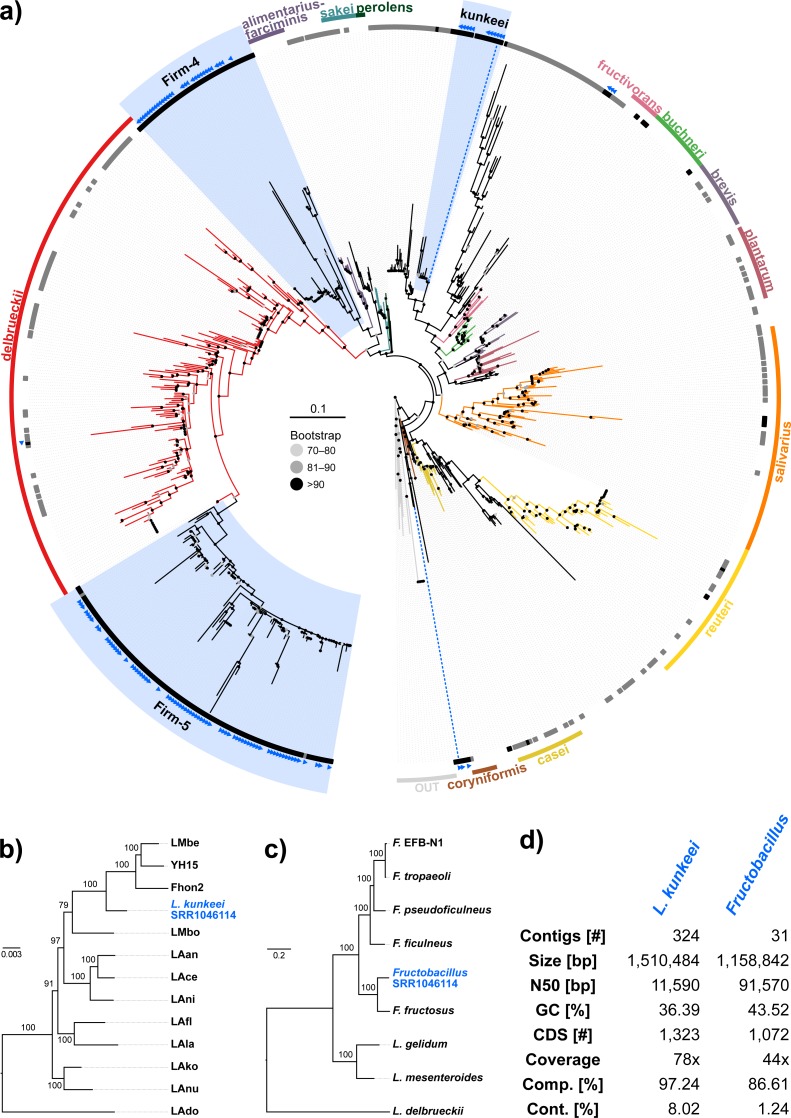
Figure 3. ‘Contamination’ from Lactobacilli in Apis short read libraries.
(A) Maximum likelihood tree of 720 16S rRNA sequences from Lactobacilli. Branch colors and the color of the outer annotation circle correspond to Lactobacillus species groups according to Felis & Dellaglio (2007). Inner circle demarks taxa found Hymenoptera (grey squares) and in corbiculate apids (honey bees and relatives, black squares). Lactobacillus sequences recovered in this study from contaminated Apis libraries are labeled with blue triangles. The Lactobacilli typically associated with honey bees (Firm-4, Firm-5, L. kunkeei) are further highlighted with a blue background color. Two dotted blue lines denote the taxa of which whole draft genomes were recovered. See text for details. An interactive version of the tree containing all node labels is available under http://www.evolgenius.info/evolview/#shared/wZcKHbwJuT. (B) Phylogeny of Lactobacillus kunkeei strains based on maximum likelihood analyses of 947 concatenated single copy orthologs (290,774 amino acid positions). Tree is rooted with Lactobacillus apinorum Fhon13 (taxon not shown). Strain names correspond to the names used in Tamarit et al. (2015; see Table S3). Blue taxon label corresponds to the L. kunkeei strain recovered from ‘contaminants’ in library SRR1046114. Bootstrap values are given on nodes. See Table S3 for sources of genomes. (B) Maximum likelihood tree of Fructobacillus (F.) and Leuconostoc (L.) species based on 435 concatenated single copy orthologs (145,069 amino acid positions). Tree is rooted with Lactobacillus delbruecki. Numbers on nodes correspond to bootstrap values. Again, blue taxon label denotes the Fructobacillus genome recovered from the ‘contaminated’ library SRR1046114. Note that the phylogenetic distance between Fructobacillus fructosus and the novel genome is similar to other between-species distances in this tree. See Table S3 for accession numbers of all genomes used for phylogenetic analysis. (D) Assembly statistics for the two novel draft genomes recovered from library SRR1046114. Abbreviations: CDS, coding sequences predicted with PROKKA; Comp. & Cont., completeness and contamination as estimated with CheckM version 1.0.6 (Parks et al., 2015) based on the number of conserved marker loci. Phylogenetic affiliations of the two strains are depicted in Figs. 3B and 3C, respectively.