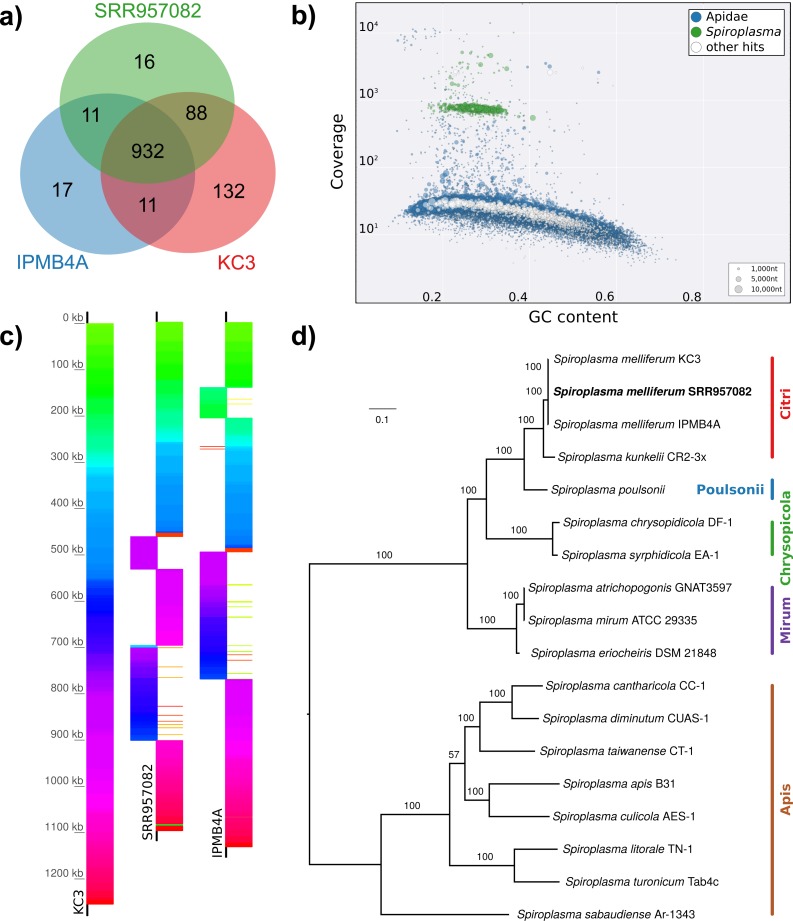
Figure 4. Characteristics of Spiroplasma melliferum. isolated from a ‘contaminated’ Apis mellifera sequencing library (SRR957082).
(A) Venn diagram illustrating the number of ortholog groups shared between the novel strain and its closest sequenced relatives IBMB4A (Lo et al., 2013) and KC3 (Alexeev et al., 2012). (B) Taxon-annotated GC-coverage plot of SRR951082 metaassembly created with Blobology. Spiroplasma and Apis contigs can be differentiated by coverage. (C) Synteny across Spiroplasma melliferum genomes. Contigs from assemblies SRR957082 and IPMB4A were ordered against KC3, the most complete of the three S. melliferum genomes. Identical colors indicate syntenic blocks of the genomes. (D) Phylogenetic relationships within the genus Spiroplasma. Maximum likelihood tree is based on 206 concatenated loci (58,950 amino acid positions), numbers on branches correspond to bootstrap values. Spiroplasma groups are highlighted with colors. The taxon label of the novel genome is highlighted in bold. Accession numbers for all taxa are listed in Table S3.