Figure 1. DAN family sequence alignment and electrostatics of Grem2.
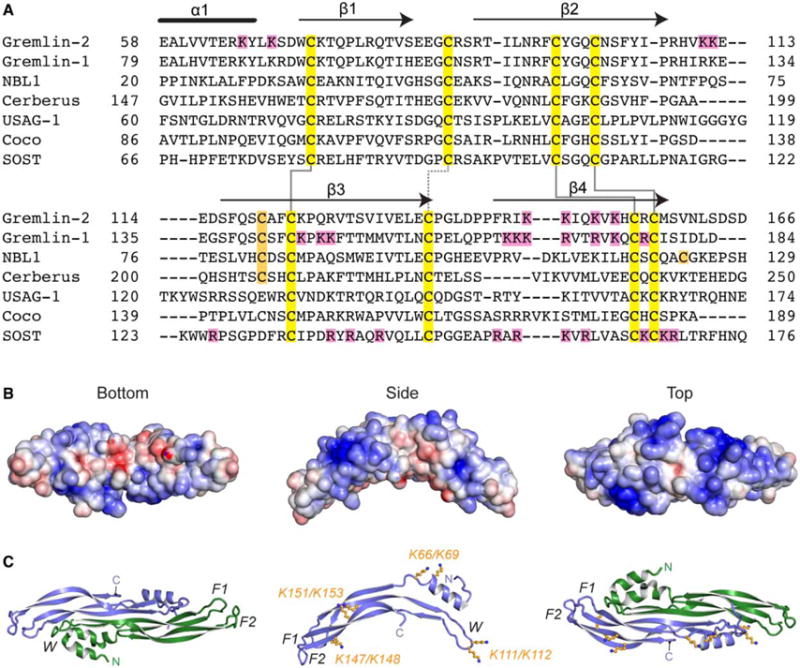
(A) Multiple sequence alignment of members from the DAN family. Alignment is based on those amino acids and ranges present in the structure of Grem2 (PDB: 4JPH). Black lines above the Grem2 sequence indicate the secondary structure. Yellow, family conserved cysteines; orange, extra cysteines; magenta, lysines in Grem2 selected for mutagenesis and those amino acids identified in Grem1 and SOST shown to be important in heparin binding. Solid black lines show disulfide bonds forming the cystine knot and the dotted black line shows the disulfide bond linking the fingers in the DAN family fold. (B and C) Structure of Grem2 shown from the bottom (concave), side, and top (convex) perspectives. (B) Electrostatic surface area predicted by APBS with blue as positive and red as negative and colored from −5 to +5 kbt/ec. (C) Ribbon representation of Grem2 dimer with chains colored green and blue. Right and left panels represent similar views as in (B). Middle panel depicts a single chain with the orange sticks depicting the lysines selected for mutagenesis studies. F1, finger 1; F2, finger 2; W, wrist region.
