Figure 7. Electrostatic surface of the Grem2–BMP2 ‘daisy-chain’ and structural comparison of different DAN family heparin/HS-binding motifs.
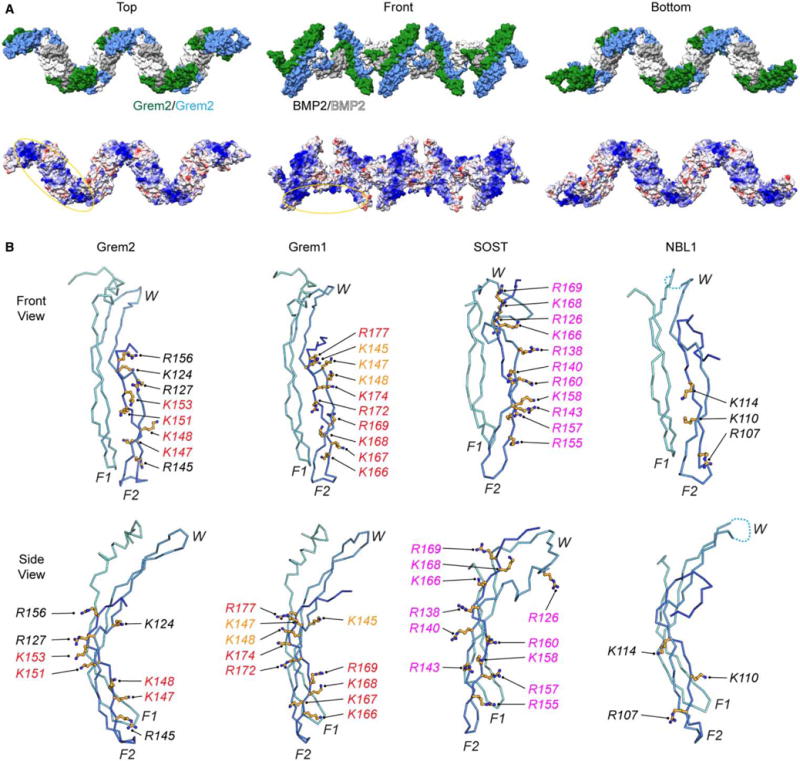
(A) Structure model of the Grem2–BMP2 polymer (‘daisy-chain’) complex made from the observed ‘daisy-chain’ complex in the Grem2–GDF5 structure (PDB: 5HK5). (Top) Surface representation of the ‘daisy-chain’ complex, with Grem2 monomers colored green and blue and BMP2 monomers colored white and gray. (Bottom) Coulombic surface coloring of the Grem2–BMP2 ‘daisy-chain’ complex as performed using UCSF Chimera. Structure is colored from −10 to +10 kbt/ec, with more negative surfaces colored in red, neutral surfaces colored in white, and positive surfaces colored in blue. Heparin/HS-binding motifs of Grem2 and BMP2 are circled. (B) Structures of Grem2 (PDB: 4JPH), Grem1 (PDB: 5AEJ), SOST (PDB: 2K8P), and NBL1 (PDB: 4X1J) shown in ribbon representation. Structures show a single monomer of each DAN family antagonist with determined or predicted heparin/HS-binding lysines or arginines shown in stick representation in orange. Orange shows amino acids in Grem1 that have been tested with moderate importance in heparin/HS binding, red shows high importance, and black shows untested amino acids that are synonymous with amino acids in other DAN family proteins shown to be important. Magenta shows amino acids in SOST that were shown to bind directly to heparin via NMR spectroscopy.
