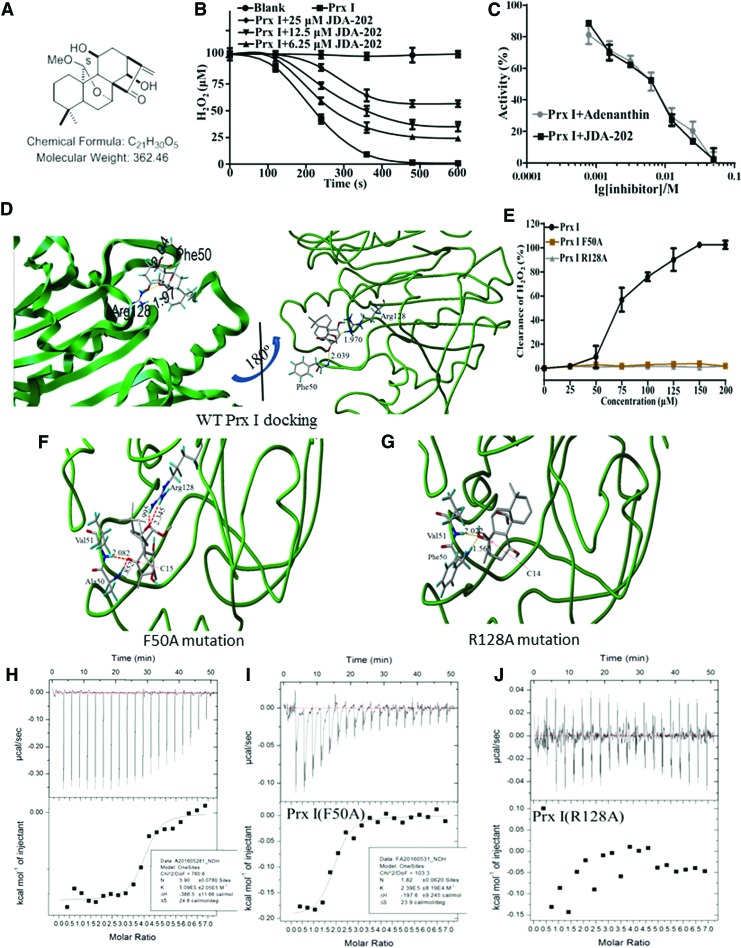
FIG. 2.
JDA-202 directly interacts with Prx I and inhibits its catalytic activity in vitro. (A) Chemical structure of JDA-202. (B) The recombinant Prx I protein was incubated with the indicated concentrations of JDA-202 for 0, 120, 240, 360, 480, and 600 s and their peroxidase activities were monitored. (C) The recombinant Prx I protein was incubated with the indicated concentrations of JDA-202 or adenanthin for 10 min, and their peroxidase activities were monitored. (D) Binding model of JDA-202 in Prx I dimerization pocket. Dotted lines represent hydrogen bonds between JDA-202 and Prx I. Right figure is the 180° northeast turn of the left figure. (E) The catalytic activities of Prx I and the two mutants (Prx I F50A and Prx I R128A) to H2O2. Data are presented as means ± SD. Three individual experiments were performed for each group. (F, G) Binding models of JDA-202 in Prx I F50A or Prx I R128A dimerization pocket. (H–J) Binding affinity of Prx I, Prx I F50A, or Prx I R128A with JDA-202. Upper graph shows raw data from one titration of JDA-202 (2 mM) into a solution containing protein (60 μM). The lower graph shows the integrated heats from the titrations of JDA-202 into a solution containing Prx I, Prx I F50A, or Prx I R128A as a function of their molar ratio. The solid lines represent the best-fit binding isotherms to the data (one set of sites model within the Origin software), (Prx I, K = 5.1 ± 2.5 × 105 M−1, ΔH = −386.5 ± 11.7 cal/mol, ΔS = 24.8 cal/mol/deg and n = 3.9 ± 0.1), and (Prx I F50A, K = 2.4 ± 0.8 × 105 M−1, ΔH = −197.6 ± 9.2 cal/mol, ΔS = 23.9 cal/mol/deg and n = 1.82 ± 0.06). H2O2, hydrogen peroxide. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars