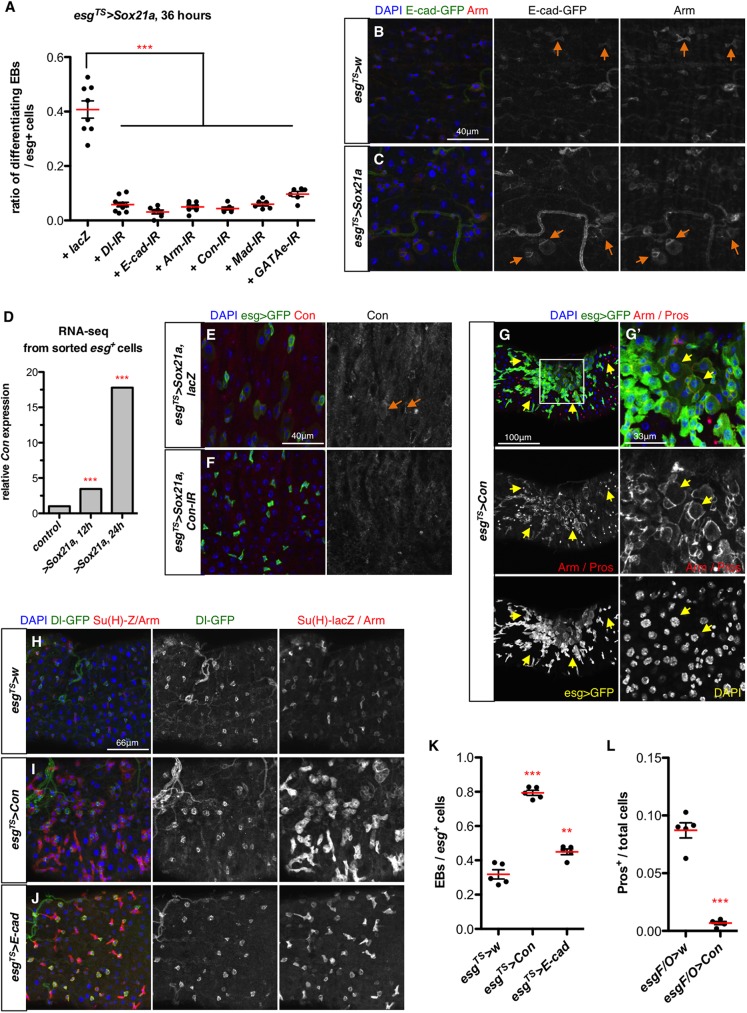
Fig 4. Cell adhesion molecules are required for Sox21a-induced differentiation.
(A) Quantification of the ratio of differentiating EBs out of all the esg>GFP+ cells in intestines of flies over-expressing Sox21a in progenitors (esgTS>Sox21a) with co-depletion of the indicated genes with RNAi for 36 hours. (B-C) Localization of E-cad (revealed with an endogenous E-cad-GFP fusion) in both control (B) and esgTS>Sox21a intestine (C). E-cad/Arm junctions are indicated by orange arrows. (D) Relative Con mRNA levels in control and Sox21a-expressing progenitors from the RNA-seq experiment described in Fig 3C. (E-F) Connectin (Con) is enriched in the progenitor contact in esgTS>Sox21a intestines (E, orange arrows) by applying an antibody against Connectin, but is not detected in esgTS>Sox21a intestines co-depleted for Connectin by RNAi (F). Progenitors (esg>GFP) and nuclei (DAPI) are shown. (G-G’) Progenitor-specific expression of Connectin using esgTS for 4 days. The region outlined in (G) is shown in (G’) with higher magnification. Note the absence of EEs (Pros+) from the region with big clusters of esg>GFP+ cells, some of which are differentiating towards EC. Some differentiating EBs are indicated with yellow arrows (G’). (H-K) representative images of midgut of control flies (H) or flies overexpressing Connectin (I) or E-cad (J) with esgTS for 4 days. ISCs express Dl-GFP (in green); EBs are marked with Su(H)-lacZ reporter (cytoplasmic, in red). Quantification of the ratio of EBs out of all the progenitors (ISC+EB) is shown in (K). (L) Quantification of the ratio of EEs (Pros+) in all the midgut cells in defined areas of the anterior midgut of flies with indicated genotype and shifted to 29°C to induce transgene expression for 7 days. Each dot represents one gut.