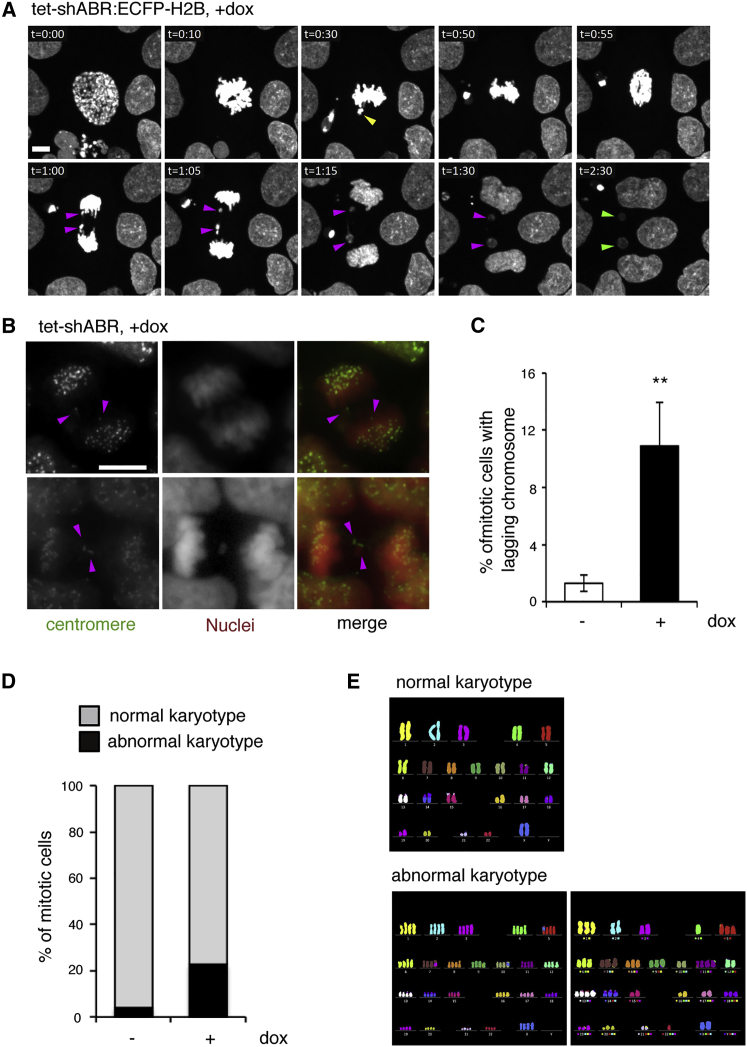
Figure 4.
Chromosome Missegregation and Aneuploidy in ABR-Depleted Cells
(A) Snapshots from Movie S3. Yellow, magenta, and green arrowheads indicate misaligned chromosomes at metaphase, lagging chromosomes at anaphase, and micronuclei in daughter cells, respectively.
(B and C) Immunostaining analyses for lagging chromosomes. Two representatives from dox-treated samples are shown in (B) (nuclei, red; centromeres, green). Magenta arrowheads indicate centromere-positive lagging chromosomes. Mitotic cells with lagging chromosomes were counted and the incidence was shown in (C). Control (n = 315) and dox-treated cells (n = 318) were analyzed.
(D) Chromosome counting analyses. Mitotic spreads were prepared using tet-shABR cells that were treated or untreated with dox for 5 days. In each sample, 50 mitotic cells were subjected to counting.
(E) Multicolor fluorescence in situ hybridization (FISH) analyses. The dox-treated sample was stained with FISH probes for each chromosome. Two independent experiments were performed, and representative examples for normal and abnormal karyotypes are shown.
The immunostaining was repeated three times with five replicates in each experiment (B and C). The mitotic spreads for chromosome counting were prepared in three separate experiments (D). Scale bars, 10 μm. Error bars in the graphs represent SD. Statistics: Student's t test (C, n = 3); ∗∗p < 0.05. See also Figure S3 and Movie S3.