Figure 1.
Generation of Thap1-Recombinant ESCs
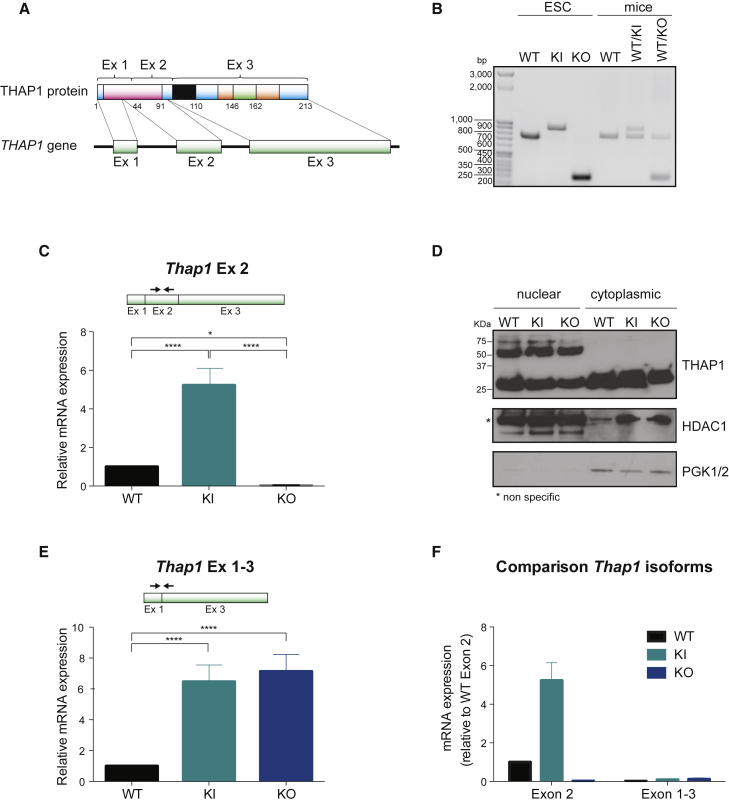
(A) Structure of Thap1 gene and its encoded protein.
(B) Genotyping of wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs by PCR and comparison with the pattern of WT, Thap1C54Y/+ (WT/KI), and Thap1ΔExon2/+ (WT/KO) heterozygote mice.
(C) Thap1 exon 2 (Thap1 Ex 2) transcript level measured by qRT-PCR in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. An ANOVA was performed which revealed a significant difference among the genotypes (F(2,23) = 91.69, p < 0.001). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of three independent experiments. ∗p < 0.05; ∗∗∗∗p < 0.001.
(D) Distribution of THAP1 protein in nuclear and cytoplasmic fraction of wild-type (WT), Thap1C54Y/C54Y (KI) and Thap1ΔExon2 ESCs (KO). Histone deacetylase 1 (HDAC1) and phosphoglycerate kinase 1/2 (PGK1/2) were used as a control of nuclear and cytoplasmic extract purity, respectively.
(E) Level of Thap1 transcript spanning exon 1 and exon 3 (Thap1 Ex 1–3) measured by qRT-PCR in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. ANOVA revealed a significant difference among the genotypes (F(2,21) = 73.85, p < 0.0001). The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of at least three independent experiments. ∗∗∗∗p < 0.001.
(F) Comparison of Thap1 isoforms by qRT-PCR in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. Data are presented as mean ± SEM.