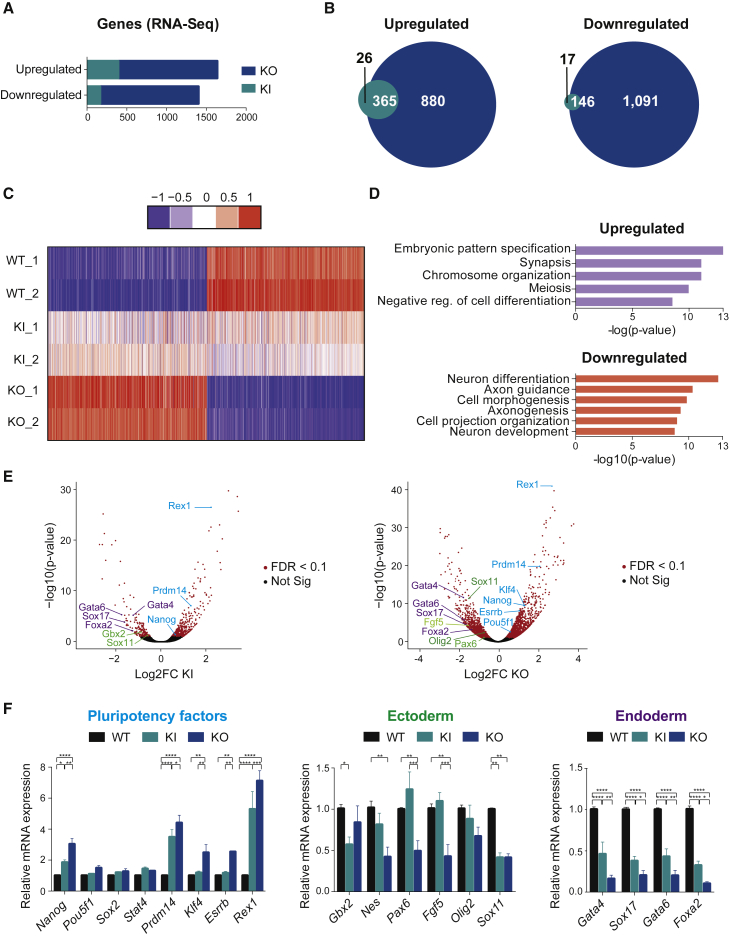
Figure 3.
THAP1 Negatively Regulates the ESC Transcriptome
(A) Bar graph illustrating the numbers of upregulated and downregulated genes in Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) compared with wild-type (WT) ESCs by RNA sequencing (RNA-Seq).
(B) Venn diagram showing the overlap of upregulated (left panel) and downregulated (right panel) genes in Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) ESCs.
(C) Heatmap illustrating the global gene expression performed in duplicates of wild-type 1 and 2 (WT_1 and WT_2), Thap1C54Y/C54Y 1 and 2 (KI_1 and KI_2), and Thap1ΔExon2 1 and 2 (KO_1 and KO_2) ESCs.
(D) Gene ontology (GO) analyses of biological processes of the top (log2 fold change 1) upregulated (upper panel) and downregulated (lower panel) genes of the Thap1ΔExon2 ESCs dataset.
(E) Volcano plots demonstrated differentially expressed genes in Thap1C54Y/C54Y (KI) (left panel) and Thap1ΔExon2 (KO) (right panel) ESCs versus WT ESCs. Significant and not significant hits are shown in red and black, respectively (false discovery rate [FDR] < 0.1). Pluripotency-associated transcripts are shown in blue. Selected ectoderm and mesoderm RNAs are depicted in green and purple, respectively.
(F) qRT-PCR analysis of selected pluripotency factors (left panel), ectoderm (middle panel), and endoderm (right panel) specification markers in WT, Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) ESCs. For pluripotency factors, two-way ANOVA was performed, revealing an interaction effect (F(14,170) = 10.92, p < 0.0001). In addition, significant differences were observed by pluripotency gene(s) (F(7,170) = 38.54, p < 0.0001) and by genotype (F(2,170) = 85.22, p < 0.0001). For ectodermal markers, two-way ANOVA was performed, revealing an interaction effect (F(10,107) = 3.178, p = 0.0013). In addition, marginal differences were observed by ectodermal gene(s) (F(5,107) = 2.144, p = 0.0657), and significant differences were observed by genotype (F(2,170) = 22.47, p < 0.0001). For endodermal markers, two-way ANOVA was performed, revealing significant differences by genotype (F(2,81) = 194.2, p < 0.0001); however, no significant differences were observed by endodermal gene(s), and there was no interaction effect. The Holm-Sidak multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for the following genes: Nanog, Prdm14, Klf4, Esrrb, Rex1, Gbx2, Nes, Pax6, Fgf5, Sox11, Gata4, Sox17, and Foxa2. Data are presented as mean ± SEM of four independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.