Figure 4.
THAP1 ChIP-Seq Analysis
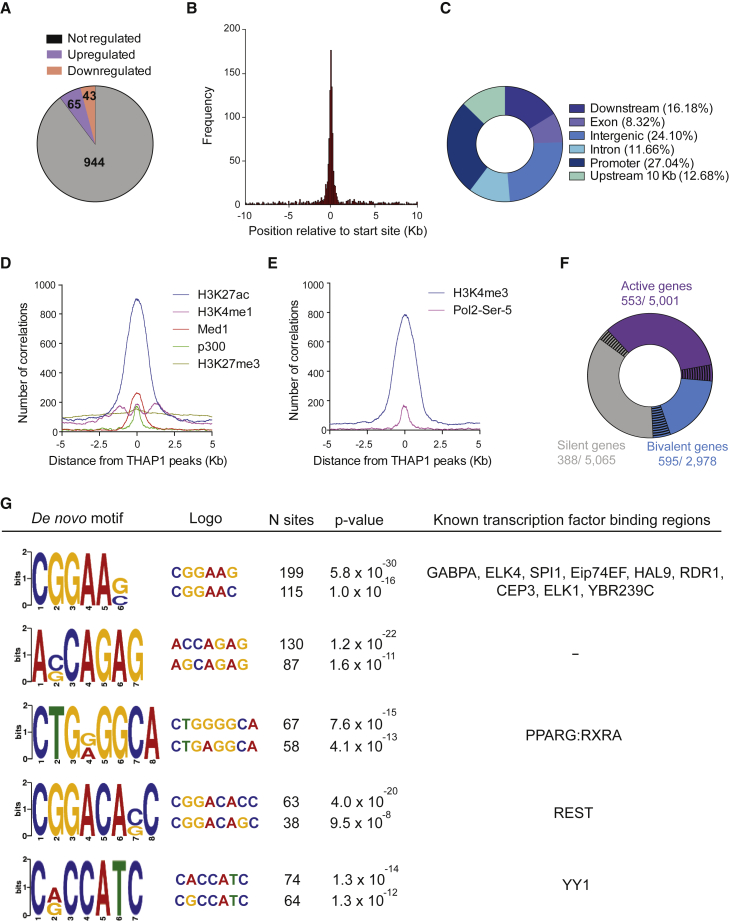
(A) Pie chart depicting the number of THAP1 only bound genes, activated and bound, and repressed and bound in Thap1ΔExon2 (KO) ESCs.
(B) Histogram showing the distribution of THAP1-binding peaks relative to the nearest TSS.
(C) Pie chart of the genomic distribution of THAP1-binding peaks including promoters (within 5 kb upstream of TSS), downstream (within 10 kb downstream of the gene), introns, exons, upstream (within 10 kb upstream of the gene), and intergenic regions.
(D) p300, MED1, H3K4me1 (characteristic of predicted enhancer), H3K27ac (active state), and H3K27me3 (repressed state) occupancy around the summit of THAP1 peaks.
(E) Pol II Ser-5P and H3K4me3 (characteristic of promoters) occupancy around the summit of THAP1 peaks.
(F) Pie chart depicting THAP1 binding sites (black lines) at active (purple), bivalent (blue), and silent (gray) genes (defined in Experimental Procedures).
(G) The top 1,000 TF ChIP-Seq peaks were used to identify THAP1 de novo motif (left panel showing the five most significant motifs) and overlapping of THAP1 binding with the indicated transcription factors (right panel). Number (N) of sites and the corresponding p values are indicated.