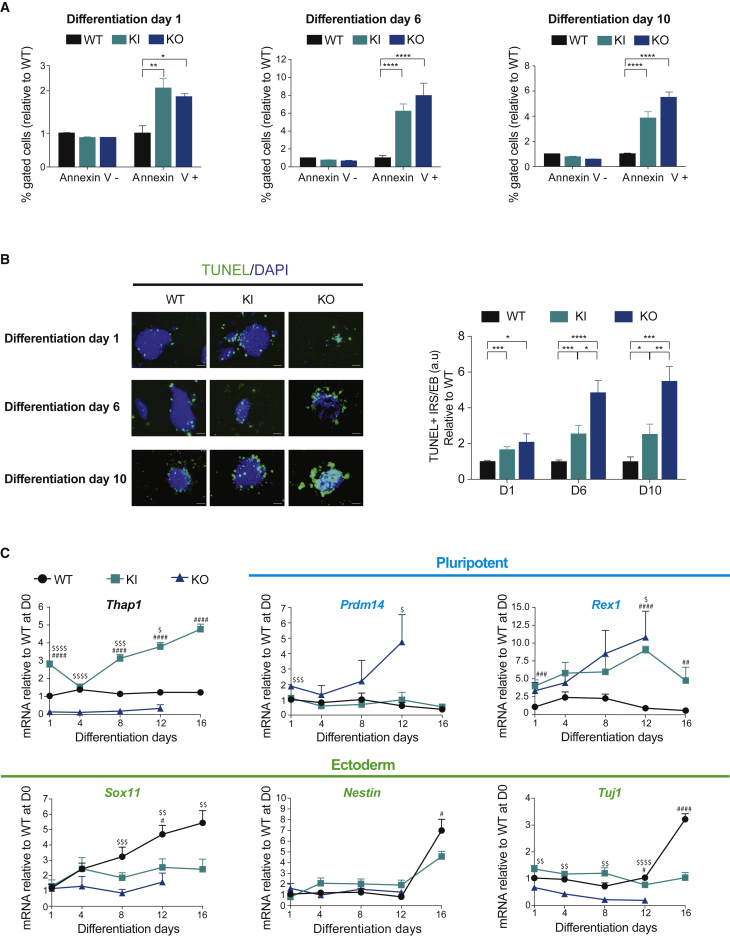
Figure 6.
Thap1C54Y/C54Y and Thap1ΔExon2 ESCs Show Abnormal EB Viability and Differentiation
(A) Percentage of live (Annexin V−) and apoptotic cells (Annexin V+) were measured over EB differentiation days 1, 6, and 10 in wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) EBs. On each day, data (mean ± SEM) are presented relative to WT = 1. The two-way ANOVA results for the annexin V assay during EB differentiation days 1, 6, and 10 can be found in Table S3. Bonferroni's multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for each differentiation day. Data are presented as mean ± SEM of three independent experiments. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗∗p < 0.001.
(B) Wild-type (WT), Thap1C54Y/C54Y (KI), and Thap1ΔExon2 (KO) EBs were examined with TUNEL on differentiation days 1, 6, and 10 (left panel; scale bar, 50 μm). Quantification of TUNEL+ EBs (right panel) was performed by scoring TUNEL-immunopositive cells (immunoreactive species [IRS+]) as a function of total EB area, normalized to WT for each differentiation day. The two-way ANOVA results for TUNEL IRS+/EB quantifications during EB differentiation days 1, 6, and 10 can be found in Table S3. Bonferroni's multiple comparisons test was performed post hoc, revealing significant differences between the genotypes for each differentiation day. Data are presented as mean ± SEM; n = 10 per group for each day, with data pooled from three independent experiments. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
(C) qRT-PCR analysis of Thap1, pluripotency markers, and ectodermal markers during EB differentiation of Thap1C54Y/C54Y (KI) and Thap1ΔExon2 (KO) ESCs at the indicated time points. Data are normalized to WT, relative to D1. The two-way ANOVA results for Thap1, pluripotency markers (Prdm14 and Rex1), and ectodermal markers (Sox11, Nestin, and Tuj1) for differentiation days 1, 6, and 10 can be found in Table S3. Bonferroni multiple comparisons test was performed post hoc, revealing significant differences between the genotypes. Data are presented as mean ± SEM of three independent experiments, relative to WT. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. # denotes differences between KI versus WT; $ denotes differences between KO versus WT.