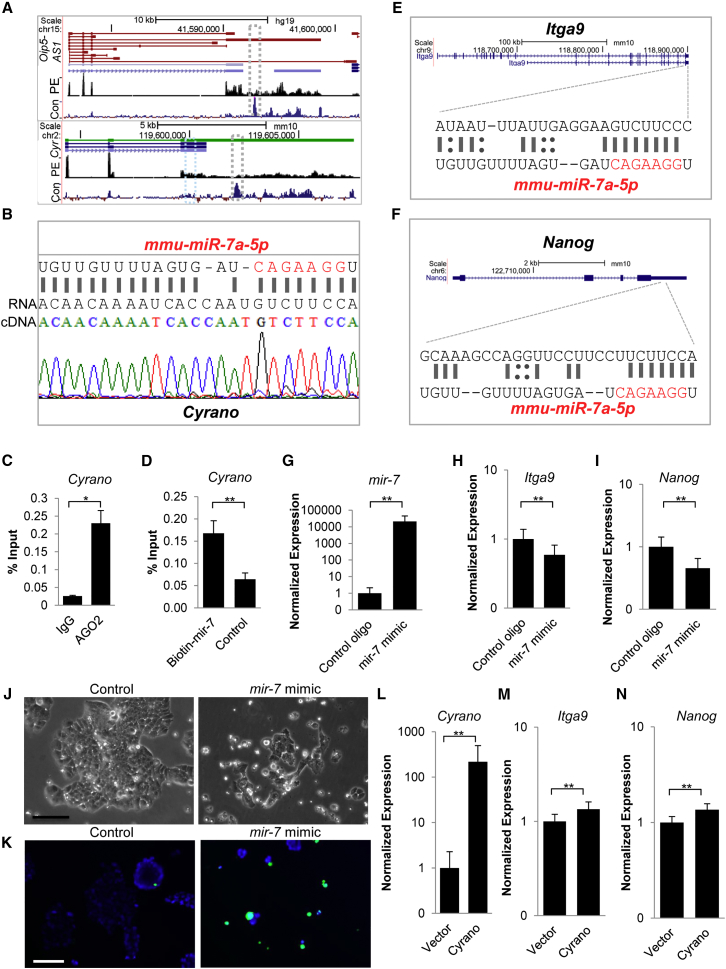
Figure 5.
Cyrano Restrains mir-7 Activity to Support ESC Self-Renewal
(A) Genome browser blots showing Oip5-AS1/Cyrano splice variants. Despite the presence of multiple splice variants of Oip5-AS1, the long variant containing the conserved mir-7 binding site (gray box; segment of this region with the mir-7 interaction site is expanded in B) is prominently expressed in human ESCs (top panel, GEO: GSE41009) and mouse ESCs (bottom panel, GEO: GSE36799). Blue box marks additional conventional mir-7 seed sequence. Con denotes conservation: Vertebrate Multiz Alignment & Conservation; PE, paired-end RNA-seq; Cyr, Cyrano.
(B) Near-complete mir-7 sequence complementarity is observed in Cyrano sequenced from ESCs.
(C) RNA immunoprecipitation indicates that Cyrano is bound by Ago2 in ESCs. Experiments were performed in triplicate with error bars representing SEM.
(D) miRNA pull-down and qPCR indicates a physical interaction between Cyrano and mir-7. Experiments were performed in triplicate with error bars representing SEM.
(E and F) The 3′ UTRs of Itga9 and Nanog contain mir-7 target sites (see Figure S5; Tables S1 and S2 for further information on target sites).
(G–N) Overexpression of mir-7 (G) results in a decrease in Itga9 (H) and Nanog levels (I), loss of ESC self-renewal maintenance (J), and increased cell death (K) in ESCs 2 days post transfection. Nuclei (blue, live; green, dead); Scale bar, 100 μm. qRT-PCR monitoring of Cyrano (L), Itga9 (M), and Nanog (N) levels with Cyrano overexpression. Error bars for qRT-PCR expression analysis represent 95% CI.
Data are from three independent experiments. ∗p < 0.05, ∗∗p < 0.01. See also Figure S5; Tables S1 and S2.