Figure 1.
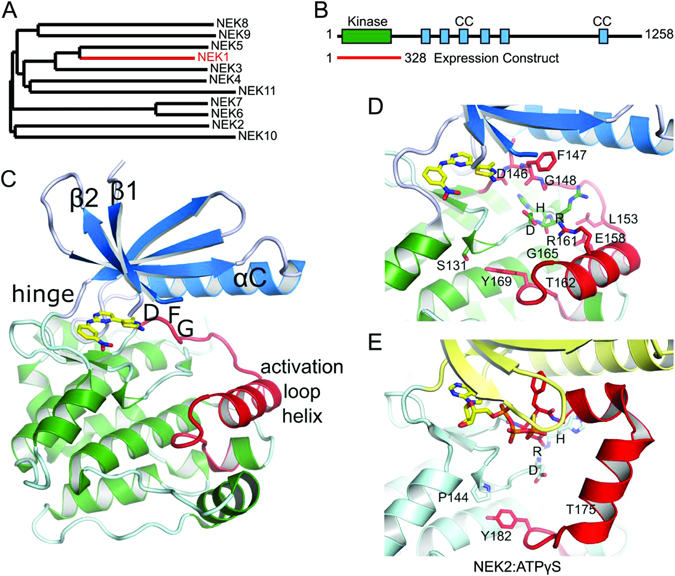
NEK1 crystal structure. (A) Phylogenetic tree of the NEK family. An alignment of the kinase domains was created using Clustal Omega75 and the phylogenetic tree drawn using TreeDyn56. (B) Domain organisation of NEK1, showing the range of the expression construct that was crystallized, and the locations of coiled-coil motifs as predicted by COILS76. (C) Structure of NEK1 protein kinase domain. The N-terminal lobe of the kinase domain (residues 1–83) is shown in blue and the C-terminal lobe in green. The activation loop (residues 146–173) is shown in red. The bound CDK2/CDK9 inhibitor 1 is shown in yellow. (D,E) Comparison of the activation loops of NEK1 (D) and CDK2 bound to ATPγS (E), based on PDB ID 2W5B77. Both kinases are in an inactive conformation with substantially different conformations of both the activation loop and the catalytically important HRD motif. Only the position of the Tyr residue at the end of the activation loop (Tyr169/Tyr182) is conserved.
