Figure 3.
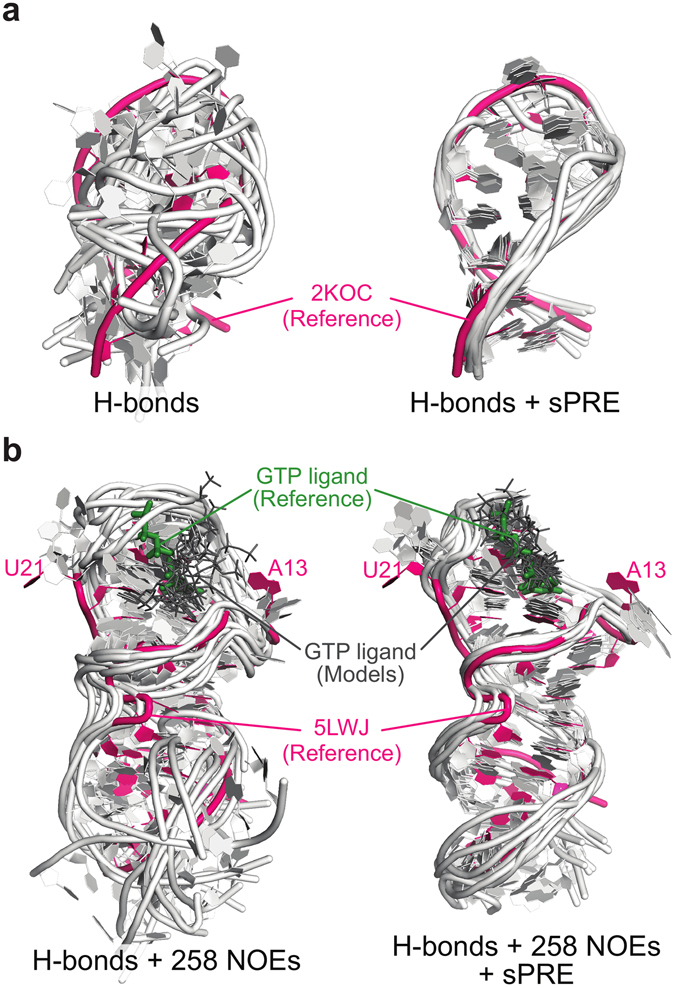
sPRE data improve structure determination of RNAs. Structural models of the UUCG tetraloop (a) and the GTP-bound GTP aptamer (b) were obtained without (left) and with (right) sPRE data using XplorNIH. The 10 best scored models (light gray) in terms of total energy were selected from a total of 200 models and aligned to the corresponding NMR structure (magenta). All restraints used in the computations are indicated below the respective models. In (b) heavy atoms of the GTP ligand are shown as sticks (Reference in green, computed models in dark gray) and the positions of the intrinsically flexible nucleotides A13 and U2140 are indicated.
