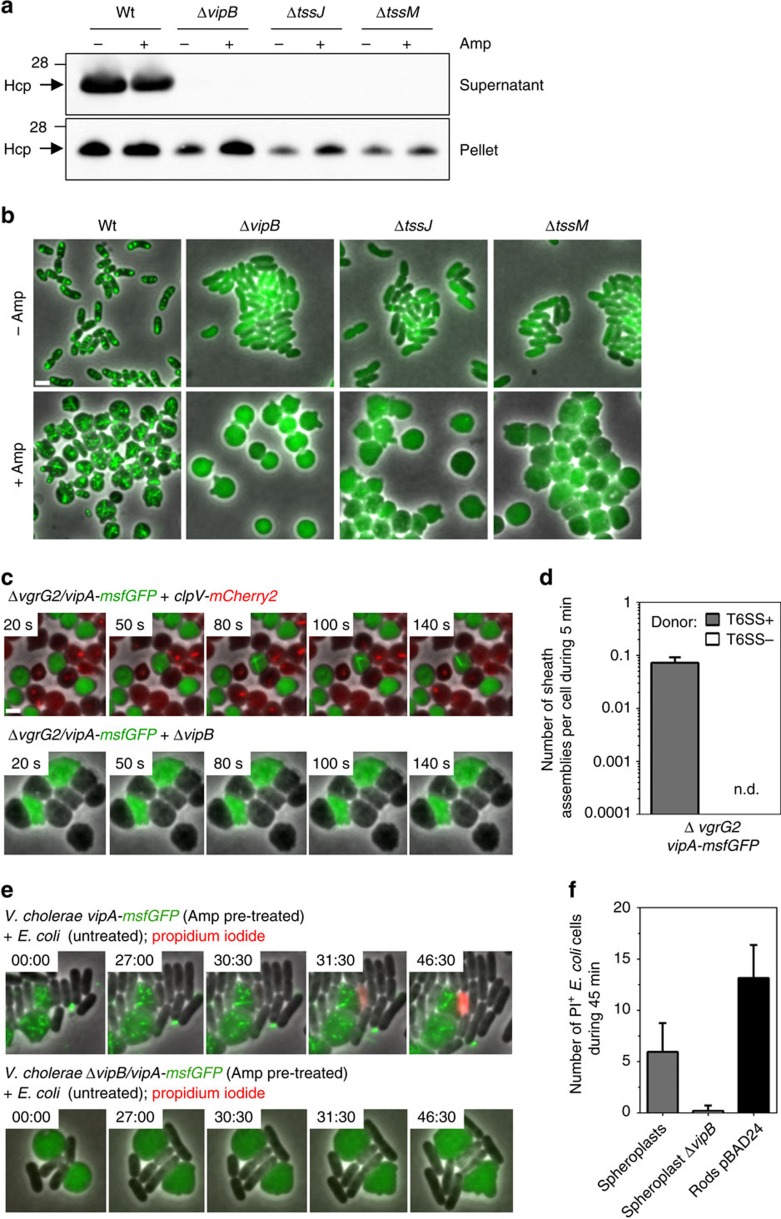
Figure 2. The T6SS appears to be functional in ampicillin-induced spheroplasts.
(a) Preinduced spheroplasts (500 μg ml−1 ampicillin for 40 min) and untreated rod-shaped cells were washed twice and inoculated in fresh medium. Presence of Hcp was detected in culture supernatant and cell pellets of indicated strains after 20 min incubation. The molecular weight is indicated on the left in kilodaltons. Full blots and Coomassie Blue stained gels are provided in Supplementary Fig. 2a. (b) Indicated strains (all vipA-msfGFP background) were monitored for sheath assembly. Representative images of untreated rod-shaped cells and spheroplasts (500 μg ml−1 ampicillin for 40 min) are shown. Scale bar, 2 μm. (c,d) VipA-msfGFP labelled recipient spheroplasts (ampicillin 500 μg ml−1, 40 min) lacking VgrG2 were co-incubated with either ClpV-mCherry2 labelled (top) or T6SS-negative (ΔvipB; bottom) donor strains. Sheath assembly in recipient cells was monitored (c) and quantified (d) in N=2,000 VipA-msfGFP cells for each mixture. See Supplementary Movie 4 for complete time-lapse series. Full fields of view are provided in Supplementary Fig. 2b. (e) VipA-msfGFP labelled T6SS positive (top) and T6SS-negative (bottom) V. cholerae spheroplasts (pretreated in ampicillin 500 μg ml−1 for 40 min, washed in LB) were co-incubated with MG1655 prey cells (grey) on agarose pads containing 100 μg ml−1 ampicillin and 1 μg ml−1 PI. (f) Number of PI+ E. coli cells was quantified from 20 fields of view (30 × 30 μm) during 45 min co-incubation with indicated V. cholerae strains. See Supplementary Movie 5 for complete time-lapse series. Full fields of view are provided in Supplementary Fig. 2c. All data were acquired from three independent biological experiments and are represented as mean±s.d. Scale bar, 1 μm.