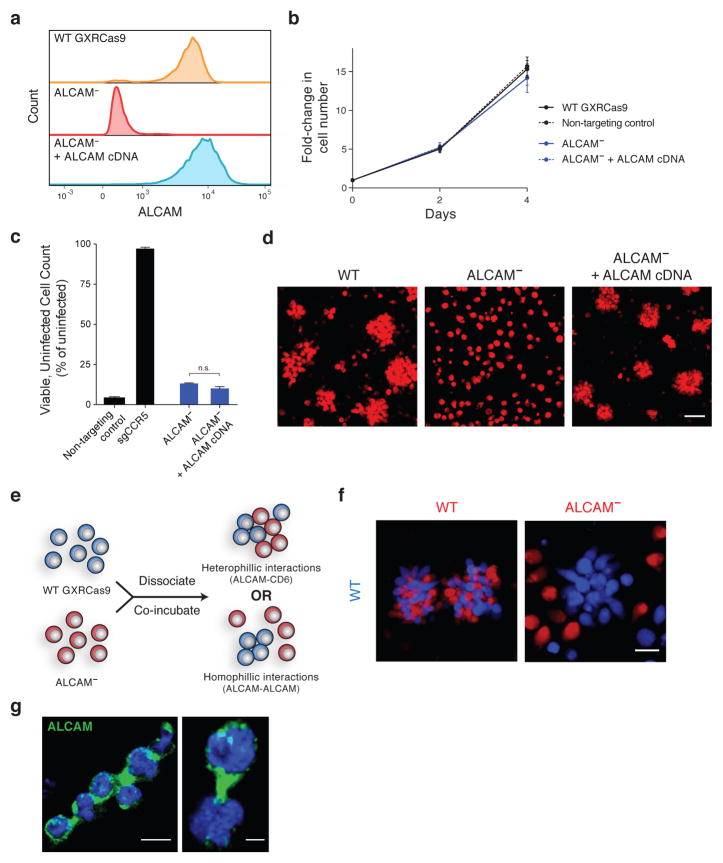
Figure 4. Homophilic ALCAM interactions are necessary for GXRCas9 cell aggregation.
(a) Flow cytometry of surface ALCAM expression in wild-type GXRCas9 cells and cells with ablated or rescued ALCAM expression.
(b) Normal proliferation of ALCAM-null cells compared to wild-type, non-targeting control, and ALCAM-rescued cells. Error bars represent standard deviation of six replicate wells.
(c) Virus challenge assay with JR-CSF demonstrating that ALCAM-null cells lack protection against HIV infection at an MOI of 1. Error bars, s.d.; triplicate wells; n.s., P=0.0308, Welch’s t-test.
(d) Confocal microscopy depicting the cellular aggregation phenotype of GXRCas9 cells with wild-type, null, or rescued ALCAM expression. Scale bar = 50 μm.
(e,f) Co-culture assay to distinguish between two models for ALCAM-mediated cell aggregation. (e) Assay design with possible outcomes. Mixed aggregates will result from the co-culture of wild-type and ALCAM-null cells if aggregates are due to heterophilic ALCAM-CD6 interactions while wild-type-only aggregates will result if aggregates are due to homophilic ALCAM-ALCAM interactions. (f) Confocal microscopy demonstrating that ALCAM-null cells are not contained within aggregates. Scale bar = 20 μm.
(g) Confocal microscopy of wild-type GXRCas9 cell aggregates for ALCAM (green) and DAPI (blue). Scale bar = 10 μm (left) and 3 μm (right).