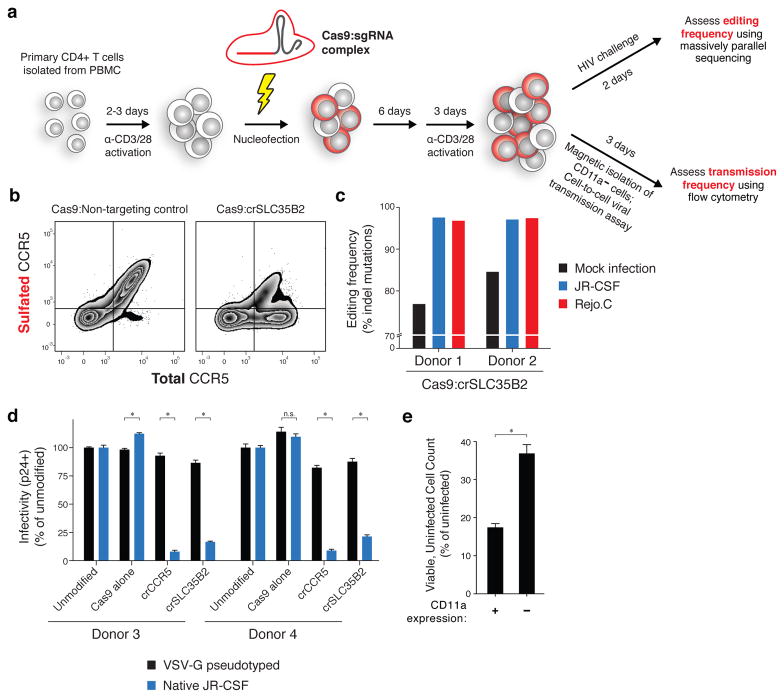
Figure 6. CRISPR-based approach for validation of HIV host dependency factors in primary human CD4+ T cells.
(a) Schematic. Primary CD4+ T cells are activated using antibodies against CD3 and CD28 and nucleofected with Cas9-ribonucleoproteins. After 6 days, cells are re-activated for 3 days and either challenged directly with HIV and sequenced at the sgRNA target site to assess mutation frequency, or purified for CD11a- cells using magnetic beads and used in a cell-to-cell HIV transmission assay.
(b) Flow cytometry of total and sulfated CCR5 surface expression in primary CD4+ T cells transfected with Cas9-RNP complexes. Cells were analyzed at the time of HIV infection.
(c) Insertion/deletion (indel) mutation frequency in primary CD4+ T cells from two donors following challenge with JR-CSF or Rejo.C, compared to a mock infection condition. See also Supplementary Fig. 4.
(d) Flow cytometry of intracellular HIV Gag in Cas9-RNP transfected primary CD4+ T cells from two additional donors. Values are normalized to un-transfected cells for each respective virus type. Entry of VSV-G pseudotyped HIV is independent of CD4 and CCR5. Error bars, s.d.; triplicate wells; * denotes P<0.01, Welch’s t-test. All * P<0.0001 except Donor 3 SLC35B2, P=0.0002. n.s., p=0.1715.
(e) Cell-to-cell HIV transmission assay in primary CD4+ T cells transfected with Cas9:crITGAL. Assay is set up as in Figure 5 except donor cells are infected 24 hours prior to co-culture and co-culture is for 2 days. Readout is by flow cytometry following intracellular staining for HIV Gag. Error bars, s.d.; triplicate wells; P=0.0036, Welch’s t-test. See also Supplementary Fig. 6.