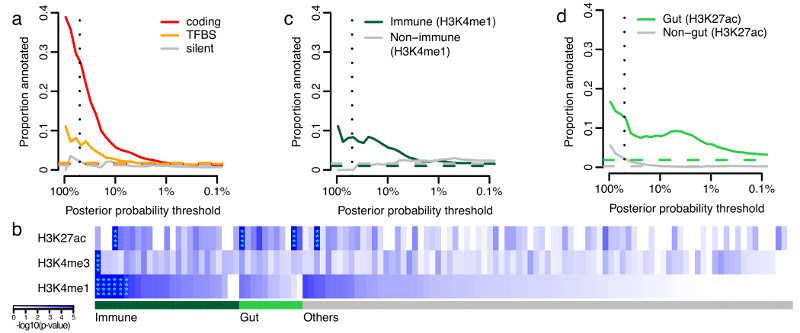
Figure 3. Functional annotation of causal variants.
a, Proportion of credible variants that are protein coding, disrupt/create transcription factor binding motif sites (TFBS) or are synonymous, sorted by posterior probability. b, Epigenetic peaks overlapping credible variants in cell and tissue types from the Roadmap Epigenomics Consortium39. Significant enrichment has been marked with asterisks. Proportion of credible variants that overlap (c) core immune peaks for H4K4me1or (d) core gut peaks for H3K27ac (Methods). In panels a, c and d, the vertical dotted lines mark 50% posterior probability and the horizontal dashed lines show the background proportions of each functional category.