Figure 1. Repertoire of non-synonymous somatic mutations of metaplastic breast cancers (MBCs) and triple-negative invasive carcinomas of no special type (IDC-NSTs).
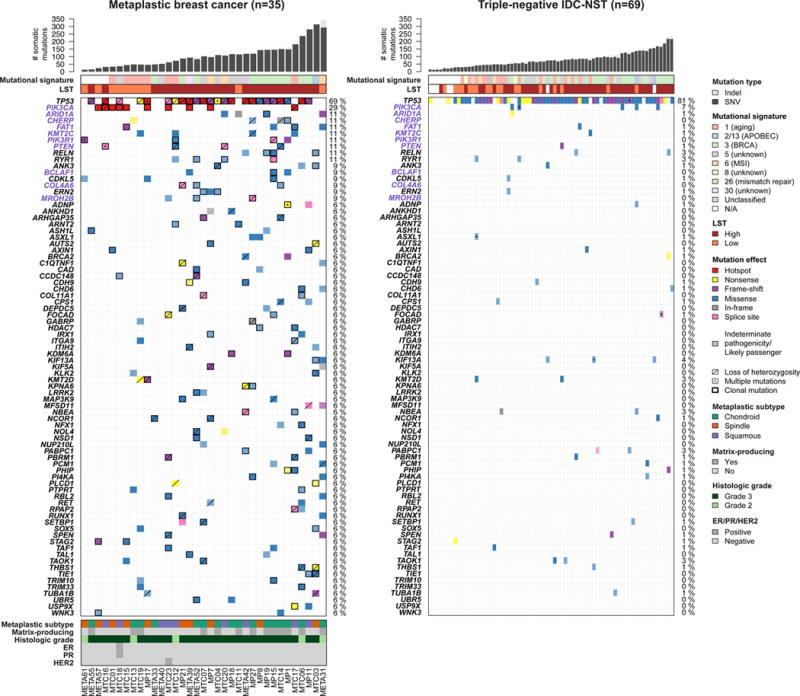
Clinico-pathologic and immunohistochemical features, and non-synonymous somatic mutations identified in 35 MBCs subjected to whole-exome sequencing (left), and in 69 triple-negative IDC-NSTs from TCGA breast cancer study (right) (9). The effects of the mutations are color-coded according to the legend, with hotspots (32) colored in red. Likely passenger mutations and mutations of indeterminate pathogenicity are marked using a hatched pattern. The presence of multiple non-synonymous mutations in the same gene is represented by an asterisk. For MBCs, the presence of loss of heterozygosity of the wild-type allele of a mutated gene is represented by a diagonal bar, and mutations found to be clonal by ABSOLUTE (24) are indicated by a black box. Genes recurrently mutated in MBCs and displaying at least one likely pathogenic mutation are presented. Gene names highlighted in purple were significantly more frequently altered in MBCs as compared to triple-negative IDC-NSTs. Percentages to the right of the mutation heatmaps indicate the percentage of cases affected by non-synonymous somatic mutations in a given gene. Bar charts (top) indicate the number of non-synonymous and synonymous somatic single nucleotide variants (SNVs) and the number of somatic insertions and deletions (indels) for each sample. The dominant mutational signatures (35, 36) were assigned using deconstructSigs (37). Large-scale transitions (LST)-high and LST-low status was determined in accordance with Popova et al. (39). TCGA, The Cancer Genome Atlas.
