Figure 3. Repertoire of non-synonymous somatic mutations affecting genes associated with the PI3K/AKT/mTOR and Wnt pathways in metaplastic breast cancers (MBCs) and triple-negative invasive carcinomas of no special type (IDC-NSTs).
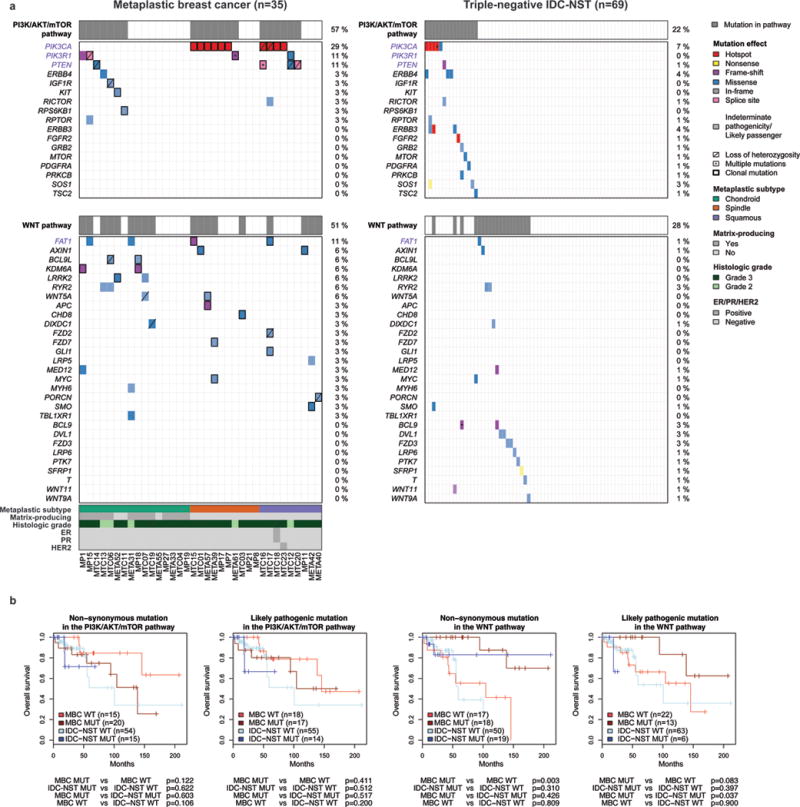
(a) Non-synonymous somatic mutations identified in 35 MBCs subjected to whole-exome sequencing and 69 triple-negative IDC-NSTs from TCGA (9) are color-coded by their effect according to the legend, with hotspots (32) colored in red. Likely passenger mutations and mutations of indeterminate pathogenicity are marked using a hatched pattern. Genes associated with the KEGG PI3K/AKT/mTOR and Gene Ontology Wnt pathways (including FAT1, see Supplementary Table S6) and mutated in at least one MBC or triple-negative IDC-NST are included and ordered in decreasing order of mutational frequency in MBCs. The presence of multiple non-synonymous mutations in the same gene is represented by an asterisk. For MBCs, the presence of loss of heterozygosity of the wild-type allele of a mutated gene is represented by a diagonal bar, and mutations found to be clonal by ABSOLUTE (24) are indicated by a black box. Percentages to the right indicate the percentage of cases affected by non-synonymous somatic mutations in a given gene. Gene names highlighted in purple were significantly more frequently altered in MBCs. (b) Overall survival of patients with MBCs or triple-negative IDC-NSTs from TCGA that harbored and did not harbor somatic non-synonymous or likely pathogenic mutations in the KEGG PI3K/AKT/mTOR and Gene Ontology Wnt pathways (including FAT1, see Supplementary Table S6) using the Kaplan–Meier method. TCGA, The Cancer Genome Atlas.
