Abstract
JIB-04, a specific inhibitor of the O2-activating, Fe-dependent histone lysine demethylases, is revealed to disrupt the binding of O2 in KDM4A/JMJD2A through a continuous O2-consumption assay, X-ray crystal structure data, and molecular docking.
Jumonji C (JmjC) domain-containing histone demethylases (HDMs) remove methyl groups from histone lysine residues through an Fe(IV)–oxo radical mechanism.1 These lysine demethylases (KDMs) belong to the larger class of α-ketoglutarate (αKG)-dependent, non-heme iron monooxygenases and play integral roles in the maintenance of genomic integrity and transcription regulation.2,3 JmjC-HDMs are considered attractive therapeutic targets as their expression is often increased in primary tumors,4 but the complex histone recognition network of these enzymes makes the design of primary substrate competitive inhibitors difficult.5,6 As a result, the vast majority of reported JmjC-HDM inhibitors are bidentate iron chelators acting as αKG competitors.7–9
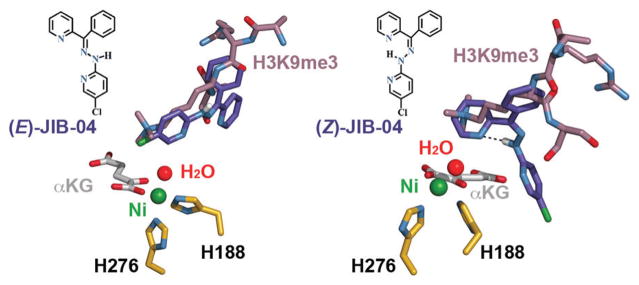
JIB-04 ((E,Z)-N-(5-chloro-pyridin-2-yl)-N′-(phenyl-pyridin-2-yl-methylene)-hydrazine) was the first reported specific inhibitor of JmjC-HDMs that is neither a competitor of αKG nor a histone peptide mimic.10,11 This pyridine hydrazone exists in two forms (Fig. 1) and displays isomer-specific inhibition with (E)-JIB-04 a more potent inhibitor than the Z-isomer. (E)-JIB-04 was found to reduce cancer growth in vivo without general toxicity, and tumour lysates of JIB-04-treated mice displayed markedly lower levels of HDM activity compared with tumour lysates of vehicle-treated mice.12
Fig. 1.
JIB-04 isomers as modelled in the KDM4A active site (PDB 2Q8C); metal-coordinating residues are shown in gold.
Among the JmjC-HDM isoforms, the pure H3K4me3 (KDM5A) and H3K9me3 demethylases (KDM4D-E) are more sensitive to JIB-04 than the H3K27 (KDM6B) and mixed H3K9/H3K36 demethylases (KDM4A-C), with reported in vitro IC50 values ranging from 0.2 to 1.1 μM. Importantly, JIB-04 does not show action against the related αKG-dependent prolyl hydroxylases and ten-eleven translocation (TET) enzymes or other chromatin-modifying enzymes such as histone deacetylases.12
In competition assays, JIB-04 displayed mixed-mode inhibition with respect to αKG and potentially competitive inhibition with respect to the primary histone substrate,12 yet no structural data were used to supplement the competition assay findings. JIB-04 shows remarkable action in promoting cell death in cancer cells and does not alter the activities of other αKG-dependent enzymes, but a thorough mechanistic understanding of the inhibitor remains to be elucidated.
Here, JIB-04’s isomer-specific inhibition of JmjC-HDMs is examined through molecular modelling studies of KDM4A. A real-time O2-consumption assay was used to probe the inhibition of KDM4A by JIB-04 at varying concentrations of O2, and JIB-04’s inhibitory strength increases in low-oxygen environments, suggesting that the hypoxic environment of the cancer cells contributes to the efficacy of JIB-04 in vivo. Finally, a 2.75 Å crystal structure of KDM4A in complex with Ni(II) was formed in the presence of JIB-04, and modelling of (E)-JIB-04 within the complex suggests that it works by disrupting the binding of O2 and histone JmjC-HDM substrates.
JIB-04 was docked into a crystal structure of KDM4A in complex with αKG and an H3K9me3 peptide (sequence ARKS) without ligand-guided assistance using OpenEye’s FRED docking program, meaning that the bound conformation of the peptide substrate was not used to guide the modelling of either isomer (Fig. 1 and 2).‡13–17 The lowest-energy poses are shown, with the aromatic moieties of (E)-JIB-04 oriented in a fashion that mimics the projection of the H3K9me3 substrate toward the metal centre in the KDM4A active site, while an intramolecular hydrogen bond between the hydrazone and pyridine (H–N···N distance of 2.8 Å) limits the protein-binding opportunities of (Z)-JIB-04.
Fig. 2.
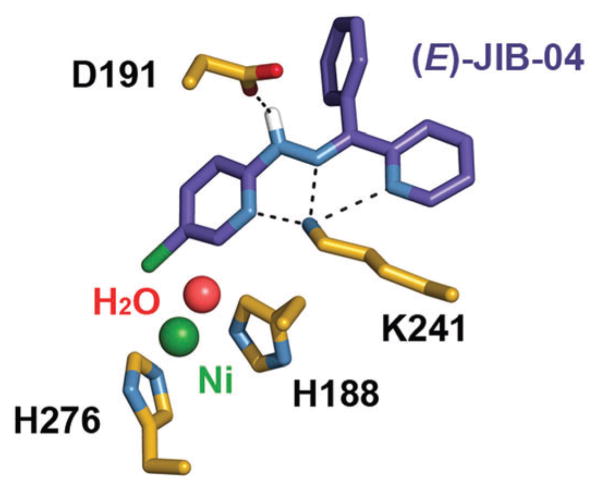
Predicted hydrogen bonding between (E)-JIB-04 and KDM4A residues, highlighted with dashes.
A closer look at the predicted binding of (E)-JIB-04 reveals hydrogen bonds between Lys 241 and the N atoms of the two pyridine and one hydrazone groups (H3N+···N distances of 2.7, 3.1, and 2.7 Å, respectively; H3N+···N distances between (Z)-JIB-04 and Lys 241 are approximately an Ångström further in each case). This hydrogen bonding is significant since Lys 241 is proposed to be essential in recruiting O2 to the active site of KDM4A.6 An additional hydrogen bond is predicted to form between the hydrazone and the carboxylate of Asp 191 (N–H···O distance of 2.4 Å). Similar to the binding of the H3K36me3 peptide in the KDM4A active site,6 we find no evidence of hydrophobic interactions to stabilize JIB-04 binding, as the nearest aromatic residue, Tyr 175, is slightly displaced and over 5 Å away.
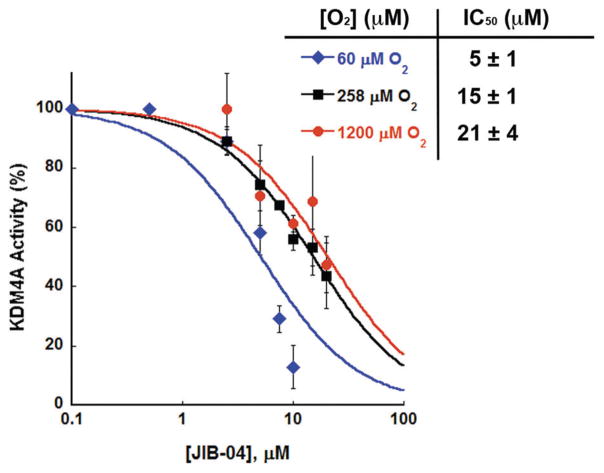
The initial report of JIB-04 found a half-maximal inhibitory concentration (IC50) of 445 nM when testing a KDM4A concentration of 206 nM, but the in vitro assay conditions were not reported in detail.§ Herein, an O2-consumption assay was used to test for inhibition of KDM4A using JIB-04 synthesized as a monohydrate.12,18 Pre-incubation of the holoenzyme with JIB-04 before the start of the reaction was necessary for notable enzyme inhibition, and this was confirmed by a MALDI-TOF mass spectrometry assay (Fig. S1, ESI†). KDM4A inhibition was screened at varying levels of O2 using a Clark oxygen electrode to track enzymatic activity in real-time (Fig. 3).19 Briefly, a solution of αKG (300 μM), ascorbic acid (500 μM), H3(7–14)K9me3 (100 μM), HEPES (pH 7.5, 50 mM), and Tween-20 (0.1%) was equilibrated under varying partial pressures of O2; outside the reaction vessel, KDM4A (2 μM, reconstituted with one equivalent Fe(II)) was incubated with JIB-04 in DMSO (0–20 μM; higher concentrations could not be tested due to the limited solubility of JIB-04). The injection of the HDM·Fe(II)·JIB-04 complex into the reaction chamber marked the start of the reaction. Enzyme inhibition was tested at three concentrations of O2: 60 μM (≈Km(O2) for KDM4A), 258 μM (air-saturated water), and 1200 μM (O2-saturated water).19 IC50 plots and values at the three concentrations of O2 are shown in Fig. 3.
Fig. 3.
Inhibition of KDM4A by JIB-04 as tested at three concentrations of O2.
With decreasing O2 concentrations we find decreasing IC50 values, and thus an increased potency of JIB-04 inhibition. While there is little difference between KDM4A’s relative activity at the three O2 concentrations with 2.5 μM JIB-04, we find substantial differences in activity at inhibitor concentrations ≥7.5 μM, with total enzyme inactivation at JIB-04 concentrations higher than 10 μM only in the low-O2 environment. Based on these data, we would expect increased inhibitor efficacy at concentrations of O2 like those found in hypoxic tissues (median pO2 ≈ 2–10 mmHg or ≈4–20 μM);20 however, KDM4A activity is difficult to accurately monitor below 60 μM O2. The nearly 3-fold difference in IC50 values between 258 and 60 μM O2 suggests that JIB-04 disrupts the binding of O2 within the KDM4A active site, while the similar IC50 values at 258 μM and 1200 μM could be expected if JIB-04 is competitive with respect to O2, since 258 μM and 1200 μM are significantly higher than the Km(O2) value for KDM4A.19 Notably, KDM4A is not the most affected target of JIB-04 since it is a mixed H3K9/H3K36 demethylase; testing the pure H3K4 and H3K9 KDMs could help to confirm that the drug exhibits a range of potencies in low- and high-O2 environments.
We were successful in crystallizing the catalytic JmjC and JmjN domains of KDM4A in complex with Ni(II) and in the presence of (E)-JIB-04, but not in the presence of (E)-JIB-04 and αKG or N-oxalylglycine (NOG).¶ The refinement of the crystallographic structure to 2.75 Å resolution revealed the electron density of the inhibitor to be unclear and so the molecular modelling of (E)-JIB-04 into the KDM4A·Ni(II) complex was performed using AutoDock Vina (ver. 1.1.2).||21 The lowest-energy pose (Fig. 4) has an apparent binding affinity of about 2 kcal mol−1 lower in energy than the most favourable poses in KDM4A·Ni(II) ·αKG (PDB 5TVR), KDM4A·Ni(II) ·NOG (PDB 2OQ7), and KDM4A·Ni(II) ·αKG·H3K9me3 (PDB 2Q8C) complexes, and approximately equal in energy to the binding affinity found for (E)-JIB-04 modelled in JARID1B/KDM5B (PDB 5A1F), the most sensitive JmjC-HDM to JIB-04 in in vitro studies (Table S1, ESI†).12,22,23 Taken together, these data indicate that (E)-JIB-04 has likely influenced the architecture of the KDM4A active site in the 2.75 Å structure.
Fig. 4.
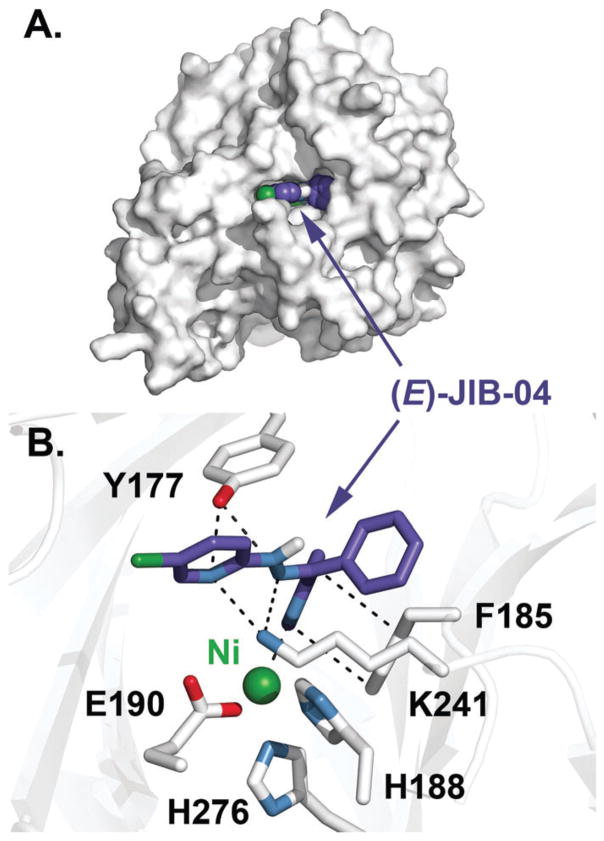
Structure of KDM4A in complex with Ni(II) and the lowest-energy (E)-JIB-04 conformer. (A) Surface view of KDM4A with a space-filling model of (E)-JIB-04. (B) Interactions of (E)-JIB-04 with KDM4A active site residues highlighted by dashes.
In the complex, (E)-JIB-04 is predicted to form two hydrogen bonds with Tyr 177 (HO···N distances of 3.1 and 3.2 Å), a residue involved in the binding of the trimethylammonium group of the enzyme’s histone substrate (Fig. 4B);5,17 this interaction is in line with earlier findings pointing to (E)-JIB-04 acting competitively with respect to the primary substrate. At least one hydrogen bond with Lys 241 forms (H3N+···N distance of 3.1 Å, with a possible second interaction at 3.7 Å), and this residue is responsible for the recruitment of O2 in KDM4A.6 Additional sources of stability in this model come from a π–π stacking interaction with Phe 185 (4 Å) and possible chelation of the metal centre by the unsubstituted pyridine nitrogen (2.7 Å), which is not possible for the Z-isomer due to its intramolecular hydrogen bond. As shown in the surface view (Fig. 4A), the hydrazone N–H group is solvent accessible and available for hydrogen bonding. While the predicted binding mode of (E)-JIB-04 in this structure is different from that found with PDB 2Q8C docking studies (Fig. 1), both models have (E)-JIB-04 interacting with Lys 241 through hydrogen bonding. This, in conjunction with the O2-consumption assay results, indicates that (E)-JIB-04 disrupts the binding of O2 in the KDM4A active site.
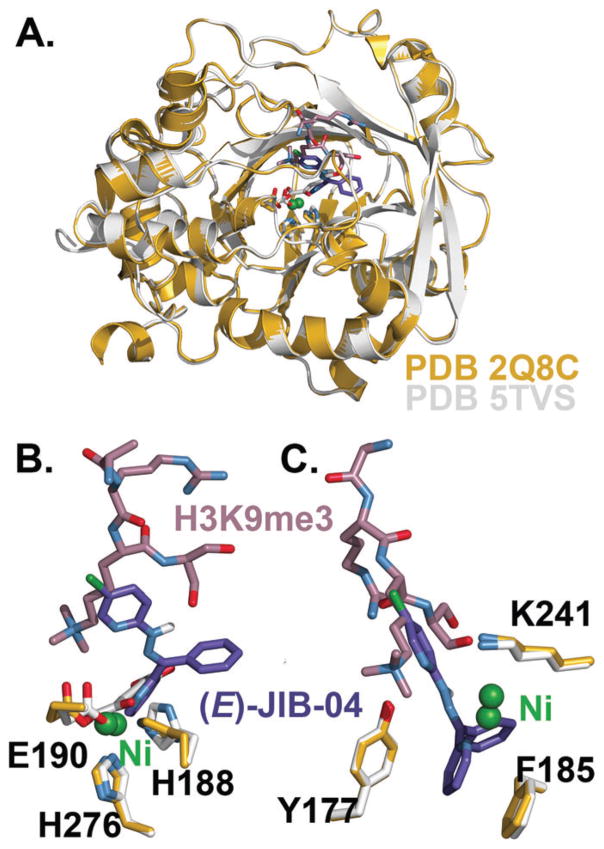
The 2.75 Å KDM4A·Ni(II) structure shows a high degree of similarity to the 2.05 Å structure KDM4A·Ni(II)·αKG·H3K9me3 (PDB 2Q8C) when the two are overlaid (Fig. 5A). However, there are notable shifts in positioning of the metal centre as well as metal-coordinating residues His 188, His 276, and Glu 190 (Fig. 5B), with Ni(II) in the 2.75 Å structure positioned about 1 Å closer to the unsubstituted pyridine nitrogen of (E)-JIB-04, which overlaps with the binding site of αKG in 2Q8C. These active site residue shifts are likely due to the presence of αKG in PDB 2Q8C rather than the presence of the histone peptide substrate, since overlaying the 2.75 Å structure with a 2.1 Å KDM4A·Ni(II)·αKG (PDB 5TVR) structure shows a similar degree of active site residue displacement (Fig. S2, ESI†). This could be an indication that the inhibitor can alter the binding of the co-substrate, since earlier studies found mixed-mode inhibition of (E)-JIB-04 with respect to αKG.12 In the histone- and O2-binding portions of the active site, only slight differences in KDM4A residues are found between the two crystal structures (Fig. 5C).
Fig. 5.
Structure comparison of PDB 2Q8C (in gold) and KDM4A·Ni(II) (in white) with (E)-JIB-04 docked. (A) Overlay of the two structures shows a high degree of structural similarity. (B) Metal centre and metal-coordinating residue displacements in the two active sites, with (E)-JIB-04 in αKG’s binding site. (C) (E)-JIB-04-binding residues show little displacement between the two structures.
In conclusion, the structural data reported here, in combination with computational docking, indicate that (E)-JIB-04 interacts with Lys 241 and Tyr 177 through hydrogen bonding, thus disrupting the binding of O2 and histone substrates in the KDM4A active site. Additionally, the inhibitor may chelate the metal centre, thereby altering the binding ability of the co-substrate αKG. Importantly, a continuous O2-consumption assay reveals that JIB-04 is a more effective inhibitor of KDM4A in low-O2 environments, a further indication that the inhibitor is competitive with respect to O2. We propose that JIB-04’s selective anticancer properties, as evidenced by reduced tumour growth without general toxicity in JIB-04-treated mice,12 are in part due to its disruption of O2-binding in JmjC-HDM active sites. While the O2 affinities of most JmjC-HDMs are unknown, the KDM4 sub-family exhibits low apparent affinities for O2, with Km(O2) values close to and above the O2 levels in well-oxygenated tissues.19 It is therefore likely that it is the low-oxygen environment of the cancer cell which allows JIB-04 to exhibit specificity of action in cancer cells over healthy cells. Our biochemical studies prompt further investigation to find how JIB-04 influences the activities of αKG-dependent HDMs in cell culture under varying tensions of O2. Overall, these studies suggest that the development of specific inhibitors for JmjC-HDMs should focus on compounds that act as competitive inhibitors with respect to more than one substrate, preferably both O2 and the histone substrate.
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available: Experimental and computational details, X-ray crystallographic data, and MALDI-TOF mass spectrometry data. See DOI: 10.1039/c6cc09882g
Ten thousand conformers of the E- and Z-isomers were generated using OMEGA following minimization by SZYBKI.24,25 Conformers were modelled into the active site using the Poisson–Boltzmann solvation method at 310 K, taking into account cavity solvation energies. All renderings were generated using PyMOL.26
In the original report, neither the concentration nor the length of H3K9me3 peptide used was specified; assuming the use of H3(7–14)K9me3, a 1: 6.5 ratio of KDM4A: H3K9me3 is calculated (400 ng KDM4A ≈ 9.1 × 10−9 mmol, 50 ng H3(7–14)K9me3 ≈ 5.9 × 10−8 mmol); the 1: 50 KDM4A: H3K9me3 ratio used here would lead to a higher IC50 value since (E)-JIB-04 is competitive with respect to the histone peptide.
KDM4A was expressed and purified as previously described, with size-exclusion chromatography (Sephadex 75) as an additional, and final, purification step.19 KDM4A (12.0 mg mL−1) was pre-incubated with 750 μM JIB-04 and 300 μM NiCl2 for 30 min before crystallization. Protein crystals were grown using the vapour diffusion method in hanging-drops of a 1: 1 mixture of protein and crystallization buffer (17.5% PEG-3350 and 0.1 M citrate pH 6.0). Diffraction data were collected at beamline 19-ID of the Argonne National Lab Advanced Photon Source. HKL30002227 was used for indexing, integration, and scaling of diffraction data. The crystal structure of KDM4A_Ni(II) was determined via a molecular replacement using PHASER2328 and the structure of the catalytic core of JMJD2A (PDB 2GP3)29 with ligands removed as the search model. Manual model building and refinement were performed in COOT30 and PHENIX,31 respectively. Data collection and refinement statistics are summarized in Table S2 (ESI†). Coordinates and structure factors for the KDM4A_Ni(II) complex have been deposited in the Protein Data Bank (5TVS).
The ligand was generated using ChemDraw 3D and was energy minimized. Twenty low-energy poses of (E)-JIB-04 were modelled in the protein complex using a grid box of 36 × 38 × 32 Å and the level of exhaustiveness set to 40.
Notes and references
- 1.Costas M, Mehn MP, Jensen MP, Que L., Jr Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik SR, Smith E, Shilatifard A. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 3.Klose RJ, Kallin EM, Zhang Y. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 4.Højfeldt JW, Agger K, Helin K. Nat Rev Drug Discovery. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 5.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BMR, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S. Proc Natl Acad Sci U S A. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang KH, King ONF, Tumber A, Woon ECY, Heightman TD, McDonough MA, Schofield CJ, Rose NR. ChemMedChem. 2011;6:759–764. doi: 10.1002/cmdc.201100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King ONF, Li XS, Sakurai M, Kawamura A, Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Beswick P. PLoS One. 2010;5:e15535. doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose NR, Ng SS, Mecinovic J, Lienard BMR, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. J Med Chem. 2008;51:7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- 10.Woon ECY, Tumber A, Kawamura A, Hillringhaus L, Ge W, Rose NR, Ma JHY, Chan MC, Walport LJ, Che KH, Ng SS, Marsden BD, Oppermann U, McDonough MA, Schofield CJ. Angew Chem, Int Ed. 2012;51:1631–1634. doi: 10.1002/anie.201107833. [DOI] [PubMed] [Google Scholar]
- 11.Lohse B, Nielsen AL, Kristensen JBL, Helgstrand C, Cloos PAC, Olsen L, Gajhede M, Clausen RP, Kristensen JL. Angew Chem, Int Ed. 2011;123:9266–9269. doi: 10.1002/anie.201101849. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, Babinski DJ, Frantz DE, Brekken RA, Quinn AM, Simeonov A, Easmon J, Martinez ED. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGaughey GB, Sheridan RP, Bayly CI, Culberson JC, Kreatsoulas C, Lindsley S, Maiorov V, Truchon JF, Cornell WD. J Chem Inf Model. 2007;47:1504–1519. doi: 10.1021/ci700052x. [DOI] [PubMed] [Google Scholar]
- 14.McGann M. J Chem Inf Model. 2011;51:578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 15.McGann M. J Comput-Aided Mol Des. 2012;26:897–906. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
- 16.Kelley BP, Brown SP, Warren GL, Muchmore SW. J Chem Inf Model. 2015;55:1771–1780. doi: 10.1021/acs.jcim.5b00142. [DOI] [PubMed] [Google Scholar]
- 17.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 18.Easmon J, Heinisch G, Pürstinger G, Langer T, Österreicher JK, Grunicke HH, Hofmann J. J Med Chem. 1997;40:4420–4425. doi: 10.1021/jm970255w. [DOI] [PubMed] [Google Scholar]
- 19.Cascella B, Mirica LM. Biochemistry. 2012;51:8699–8701. doi: 10.1021/bi3012466. [DOI] [PubMed] [Google Scholar]
- 20.Vaupel P, Mayer A, Höckel M. Methods Enzymol. 2004;381:335–354. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- 21.Trott O, Olson A. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, Strain-Damerell C, Gileadi C, Philpott M, Burgess-Brown N, Wu N, Kopec J, Nuzzi A, Steuber H, Egner U, Badock V, Munro S, LaThangue NB, Westaway S, Brown J, Athanasou N, Prinjha R, Brennan PE, Oppermann U. Nat Chem Biol. 2016;12:539–545. doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- 24.Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT. J Chem Inf Model. 2010;50:572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halgren TA. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 26.The PyMOL Molecular Graphics System, Version 1.8. Schrodinger, LLC; 2015. [Google Scholar]
- 27.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, Hagman J, Hansen K, Shi Y, Zhang G. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.