Fig. 2.
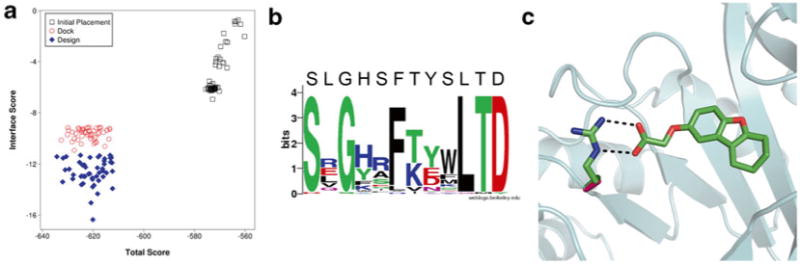
Protein/ligand interface design with RosettaLigand. (a) Comparison in improvements in Interface Score and Total Score for top models from an initial placement, docking without sequence design, and docking with design. (b) Sequence logo of mutation sites among the top models from a round of interface design [43]. For most positions, the consensus sequence resembles the native sequence. Amino acids with sidechains that directly interact with the ligand show a high prevalence to mutation as seen in the positions with decreased consensus. (c) Example of a typical mutation introduced by RosettaLigand. The protein structure is represented in cartoon (cyan). The native alanine (pink) is mutated to an arginine residue (green) to match ionic interactions with the negatively charged ligand (green). Image generated in PyMol [44]
