Figure 2.
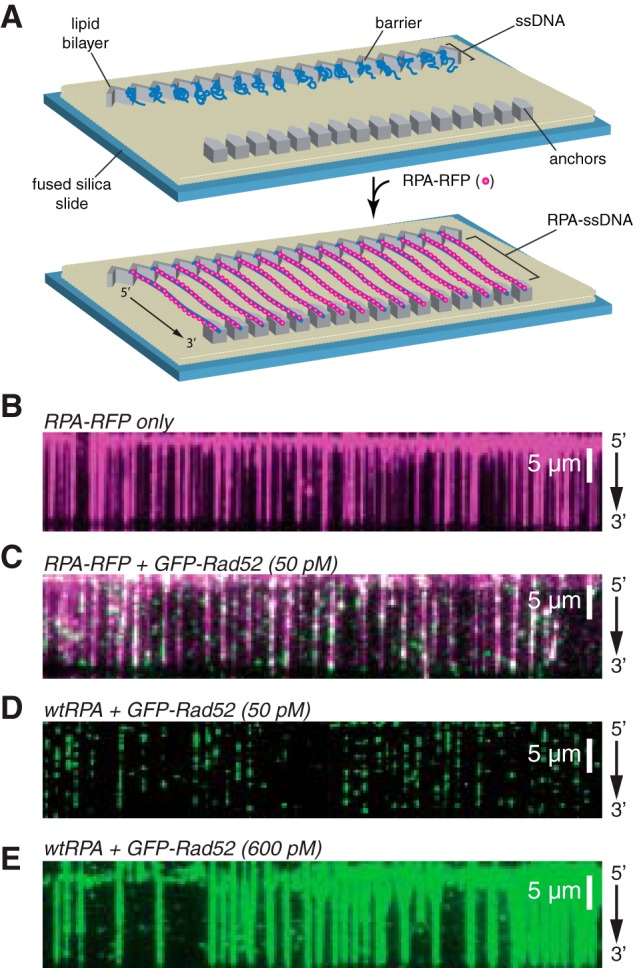
ssDNA curtain assay for RPA and RAD52 binding. A, schematic of double-tethered ssDNA curtains showing the nanofabricated patterns on the surface of a fused silica microscope slide. All hRPA-ssDNA molecules are anchored at both ends. 5′ anchoring is achieved with biotin-streptavidin-biotin linkage to the modified lipids in the bilayer and aligned at the zig-zag shaped chromium barriers. 3′ is anchored through nonspecific binding to the exposed chromium pedestals. B, wide-field image of RPA-RFP (magenta) bound to ssDNA in the absence of RAD52. C, wide-field image of RPA-RFP (magenta) curtain in the presence of 50 pm GFP-RAD52 (green). D, wide-field image of GFP-RAD52 (50 pm; green) bound to an wtRPA-ssDNA curtain (unlabeled). E, wide-field image of GFP-RAD52 (600 pm; green) bound to an wtRPA-ssDNA curtain (unlabeled).
