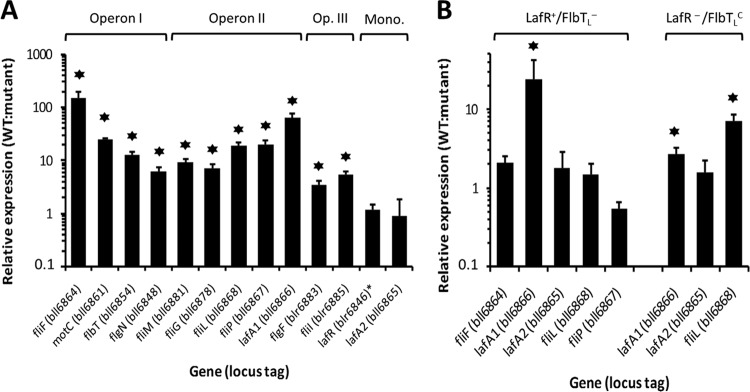
FIG 3.
Effects of mutations in lafR and flbTL on the mRNA accumulation of selected lateral-flagellar genes. (A) Transcription expression level in the wild-type strain relative to that of the lafR::Km mutant plus the standard deviations (SD), as determined by qRT-PCR from at least three independent biological replicas for the indicated genes (locus tags), the latter being representative of the different transcriptional units. Mono., monocistronic transcripts. The relative expression of lafR was evaluated with the primers indicated in Fig. S2B in the supplemental material, which amplify the 5′ end of lafR both in the wild-type and in the lafR::Km mutant. Stars, statistically significant differences (P < 0.05) from a threshold interval of 0.5 to 2.0 according to the Student t test. (B) Transcription expression level in the wild-type strain relative to that of the ΔflbTL strain (LafR+/FlbTL−, left) or that of lafR::Km carrying the plasmid pFAJ::flbTL (LafR−/FlbTLC, right) ± the SD, as determined by qRT-PCR from at least three independent biological replicas for the indicated genes, the latter having been selected to indicate the differential influence of flbTL on lafA1 expression. Stars, statistically significant differences (P < 0.05) from a threshold interval of 0.5 to 2.0 according to the Student t test. For the sake of simplicity, the L subscripts in the figure have been omitted from the name of each locus.