ABSTRACT
In mutually beneficial and pathogenic symbiotic associations, microbes must adapt to the host environment for optimal fitness. Both within an individual host and during transmission between hosts, microbes are exposed to temporal and spatial variation in environmental conditions. The phenomenon of phenotypic variation, in which different subpopulations of cells express distinctive and potentially adaptive characteristics, can contribute to microbial adaptation to a lifestyle that includes rapidly changing environments. The environments experienced by a symbiotic microbe during its life history can be erratic or predictable, and each can impact the evolution of adaptive responses. In particular, the predictability of a rhythmic or cyclical series of environments may promote the evolution of signal transduction cascades that allow preadaptive responses to environments that are likely to be encountered in the future, a phenomenon known as adaptive prediction. In this review, we summarize environmental variations known to occur in some well-studied models of symbiosis and how these may contribute to the evolution of microbial population heterogeneity and anticipatory behavior. We provide details about the symbiosis between Xenorhabdus bacteria and Steinernema nematodes as a model to investigate the concept of environmental adaptation and adaptive prediction in a microbial symbiosis.
KEYWORDS: adaptive response, Lrp, Steinernema, Xenorhabdus, symbiosis
INTRODUCTION
Microbial symbiotic associations, which are pervasive, can be beneficial (mutualistic), neutral (commensal), or harmful (pathogenic) to the host animal. Within symbioses, microbes can exploit the host for space and nutrients or as a vector for dissemination to other environments. Both within and between hosts, microbes experience changing environmental conditions to which they must adapt for optimal fitness and for maintenance of symbiotic associations. For instance, mutualistic and pathogenic symbionts that are acquired by their hosts each new generation (horizontally transmitted) experience transitions between host-associated and free-living states or among various hosts (e.g., mammals) and the vectors that transmit them (e.g., insects). These transitions can be associated with changes in key environmental parameters such as temperature, pH, and host immune factors. Similarly, an individual microbe that exclusively resides within a single host, or is passed directly to progeny through the parent (vertical transmission), can encounter dramatic shifts in environmental conditions (e.g., nutrient availability, ion concentrations, osmotic and oxidative stress) in response to host diet or hormonal shifts. For example, the mammalian gastrointestinal (GI) tract comprises numerous niches with variations in levels of sugars, pH, and metals.
To a microbe of a few microns, these variations over millimeter or centimeter scales can represent drastic environmental changes. Further, transitions among these niches can follow a predetermined pattern. For example, the process of enteropathogen infection has sequentially ordered steps: loose association with the host mucosal surface, induction of virulence factors and toxins, intimate attachment, and invasion (reviewed in reference 1). Mutualistic symbionts also undergo regimented infection processes during transmission to new hosts. In several well-described examples of horizontal transmission, such as that between the bacterium Xenorhabdus nematophila and its mutualistic host Steinernema carpocapsae, transmission is initiated by attachment to particular tissues, followed by a selective colonization bottleneck and movement of the selected symbiont to a specialized host niche (2–4).
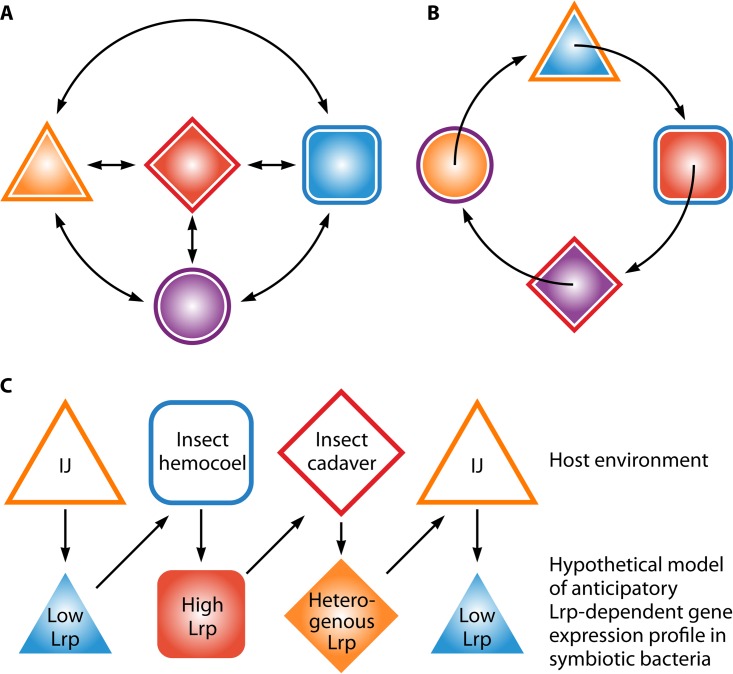
Spatial and temporal environmental shifts within symbioses can be roughly categorized into two types: erratic and predictable (Fig. 1). In the former, the symbiont, while it is regularly exposed to a finite number of different environments, does not encounter these environments in any predictable order (Fig. 1A) (5, 6). In predictable shifts, the environments experienced by a microbe during its life history fluctuate in a temporally ordered manner (Fig. 1B) (7, 8). For instance, microbes might experience such life histories when they are obligately transmitted by a vector between primary hosts (9), or within a single host due to the host's temporal rhythms, which can be entrained (circadian/circannual) or induced (diel) by light-dark periods (10).
FIG 1.
Responsive and anticipatory adaptations resulting from random and ordered environmental exposure. (A) For an organism that encounters the multiple environments (represented by outer lines), but in a random, unpredictable manner, adaptive gene expression (represented by inner shapes) occurs in response to cues or is selected by conditions encountered within the current environment (e.g., the outer and inner colors of each symbol match). (B) Temporal progression through a cyclic series of predictable environments (outer line colors: orange triangle, blue square, red diamond, and purple circle) can select for the evolution of adaptive prediction during which the cellular program of gene expression (inner shape colors: orange circle, blue triangle, red square, and purple diamond) is preadapted for the next environment. Anticipatory or preadaptive responses can occur in response to environmental cues. For instance, the orange environment (triangle) triggers a change in gene expression that is adaptive to blue environment (square) or that occurs in response to endogenous molecular clocks. (C) A working model of Lrp-dependent adaptive prediction as it may occur for Xenorhabdus bacteria, which encounter predictable stages of host interactions. Open symbols in the top row represent environments; closed symbols in the bottom row represent gene expression profiles. The color scheme represents traits being expressed in response to the current environment that would benefit fitness in the future (adaptive prediction). We hypothesize that the Lrp-dependent phenotypic switch in X. nematophila plays a role in such (pre)adaptive behavior of the symbiotic bacteria in alternating host environments.
In either predictable or erratic environmental life histories, many microbes have evolved an adaptive behavior known as phenotypic variation, a process by which individual cells within a population express distinct phenotypes that each can confer an adaptive advantage in a particular environment. The presence of more than one phenotype and the ability of microbes to switch among them provide better fitness for the microbial population as a whole. The process of phenotypic variation can be stochastic, occurring randomly within a subpopulation, or induced, in a process in which an environmental condition triggers the switching among variant phenotypes. Also, phenotypic variants that are adapted to the prevailing environment can be induced or selected by that environment (11).
In many pathogenic bacteria, phenotypic variation is thought to facilitate the transition between free-living and host-associated states. Erratic environmental fluctuations (Fig. 1A) may select for symbionts with phenotypes that switch stochastically among potential states, such that at any given time, a subpopulation of bacteria may be expressing adaptive traits appropriate to that environment, either in or out of the host. At the same time, a subset of the population may be expressing traits that would be advantageous in some future, unpredictable environment (12). Thus, a population already exists in rapidly changing environments that is preadapted to the new environment. For example, Vibrio cholerae bacteria frequently but unpredictably transit between two environments, namely, the mammalian host and the aquatic environment, which may include biofilm formation on the surfaces of invertebrates, such as copepods. Attachment to mammalian host mucosal surfaces during infection requires low levels of c-di-GMP that are necessary for virulence gene expression (13). In contrast, bacterial persistence in the aquatic environment requires high levels of c-di-GMP to ensure biofilm formation but would inversely regulate virulence genes (14). V. cholerae bacteria exhibit population heterogeneity with respect to these traits during a transient period after release from a mammalian host, called the “short-term persistence” stage. During this period in particular, the upcoming environment is erratic, since the next host or condition is unpredictable. ToxT, the master regulator of virulence, controls population heterogeneity, which results in a small subpopulation of bacteria expressing virulence genes that provide an adaptive benefit if the next environment is a new mammalian host. Concurrently, the majority virulence-off subpopulation is proposed to better adapt to biofilm formation and long-term persistence in the aquatic environment (15, 16).
Microbes that have evolved under conditions of a predictable fluctuation life history (Fig. 1B) may have the capacity to interpret prevailing conditions to anticipate an upcoming environment. Such anticipation would allow the microbe to regulate genes in a temporal order to preinduce gene profiles that are optimal for success in the predicted future environment. Experimental support for this adaptive prediction theory has been presented for Escherichia coli bacteria occupying the mammalian gastrointestinal tract, which may provide opportunities for microbes to predict and preadapt to the future host niches based on the current environmental stimuli (7, 8). For example, while passing through the GI tract, E. coli bacteria are reproducibly exposed first to lactose (signal 1, or S1) and then to maltose (signal 2, or S2). In laboratory experiments, the authors of one study showed that under conditions of exposure to lactose (S1), E. coli induces expression of a lactose gene promoter (response 1, or R1) and, to a lesser extent, a maltose gene promoter (response 2, or R2), prior to exposure to maltose (S2). This response provides an adaptive benefit for growth on maltose (S2) (7). The data indicated that the regulatory network is specifically anticipatory, such that the first signal can induce a response to an upcoming environment but not the other way around. More importantly, the authors showed that evolution in the absence of the lactose/maltose temporal link led to a weakening of this asymmetric regulation. The results of that study suggest that predictable temporal changes in the host environments can select for anticipatory behaviors in symbiotic microbes.
Microbes may experience predictable changes in environment, even if they occupy a single host niche, due to predictable rhythmic oscillations (e.g., diel, circadian, seasonal). Even though animal rhythmicity has been investigated for decades as a mechanism for anticipatory behavior, how rhythmic changes in the host environments impact the host-associated microbial community has attracted attention only recently. (For detailed examples of circadian rhythms in both mutualistic and pathogenic associations, see the review in reference 10.) Daily (circadian/diel) or seasonal alterations of animal physiology cause predictable changes to the microenvironments experienced by host-associated symbionts. For example, immunity within species as diverse as fruit fly to human is subject to circadian control, and the outcome of infection can depend on the time of day (17, 18).
Host rhythms could also cause a predictable pattern in nutrient availability for the symbiotic microbes. For instance, the light organ of the Euprymna scolopes squid, which houses the light-producing bacterial symbiont Vibrio fischeri, undergoes diel rhythmic morphological and physiological changes that have direct impacts on symbionts. At dawn, the adult squid expels the contents of the light organ, including the bulk of the symbiont population (19), and the light organ epithelium undergoes morphological changes that alter local environment (20, 21). Such host environmental changes are synchronized with symbiotic bacterial transcription profiles to express glycerol metabolism genes that support symbiont growth on host-derived glycerol substrates during the day (22). Based on transcriptomic and other data, it appears that bacterial growth in turn initiates a chemical dialog between host and microbe that allows each to adapt in anticipation of nightfall. First, the growing bacterial population induces quorum sensing and bioluminescence production a few hours before dusk, prior to the need for light emission. In the meantime, the colonization of symbionts causes host hemocyte migration into the crypts of the light organ, where they lyse and release chitin for chitin catabolism among the bacterial symbionts (23). Chitin metabolism acidifies the host tissue and further induces bacterial acid tolerance response and intensifies the bacterial luminescence production right after dusk, facilitating the host nocturnal predation (24). The symbiont microbe-associated molecular patterns (MAMPs) and luminescence also ensure the expression of host cryptochrome protein in the light organ, which is proposed to regulate host circadian behavior (25).
The examples described above indicate that host-symbiont interactions are intimately entwined with rhythmic behaviors. An open question remains as to whether evolution of symbionts in a predictable environmental regime, such as that caused by rhythmicity, has led to bacterial adaptive prediction in any of these systems. The symbiosis between Xenorhabdus bacteria and their invertebrate hosts, Steinernema nematodes and insects, may offer a particularly amenable system to investigate this issue, since it is experimentally tractable and, as described in more detail below, the temporal order of host environments encountered by Xenorhabdus is predictable (Fig. 1C).
ADAPTIVE RESPONSES IN XENORHABDUS BACTERIAL SYMBIONTS OF STEINERNEMA NEMATODES
Xenorhabdus is an insect pathogen transmitted between hosts by virtue of its ability to colonize the intestine of soil-dwelling entomopathogenic Steinernema nematodes. The infective juvenile (IJ) stage nematode carries and releases bacteria into the body cavity (hemocoel) of insects, and this infection results in rapid insect death. As part of their mutually beneficial relationship, the bacteria and nematodes both use the insect cadaver as the nutrient source to support reproduction. Once these nutrients are depleted, Xenorhabdus bacteria colonize the IJ transmission stage of the nematode, which migrates to the soil to repeat the life cycle. The general stages of the Xenorhabdus life cycle, insect infection, reproduction within the cadaver, and colonization of the IJ for transmission to another insect host (26), represent a predictable series of host environments encountered during the Xenorhabdus life history (Fig. 1C). The evolutionary success of Xenorhabdus depends on its adaptive responses to these environments, through expression of genes that encode pathogenic (toward insects) and mutualistic (toward nematodes) activities, and to the different tissues and host responses encountered (26, 27). We describe these temporally ordered environments in more detail below.
FROM THE IJ TO THE INSECT
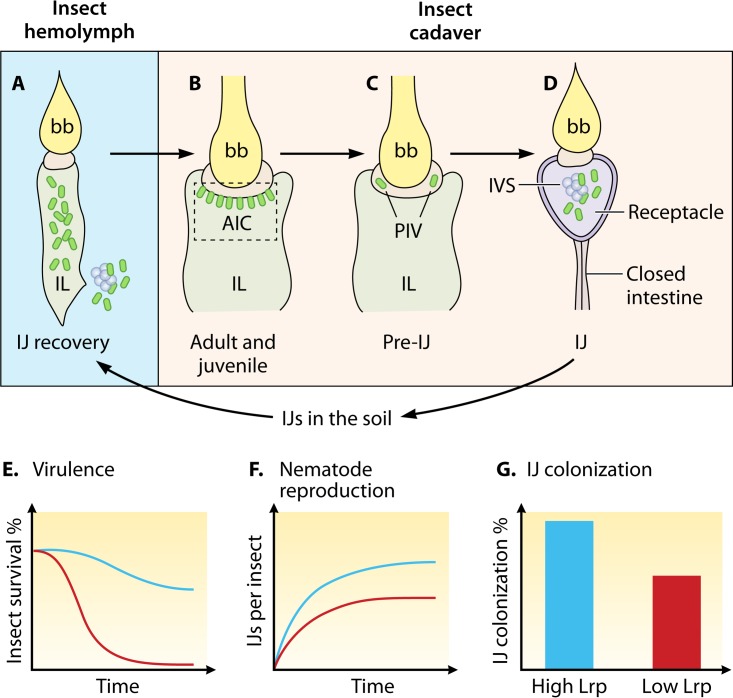
Tens to hundreds of Xenorhabdus bacterial cells colonize and persist for months in the relatively nutrient-poor intestinal receptacle of soil-dwelling IJ nematodes (Fig. 2D) (28–32). IJs can persist in the soil for many months (33–36), and in the IJ receptacle, Xenorhabdus likely expresses traits that support its long-term stationary-phase survival. IJ infection of an insect (Fig. 2A) is an obligate step in the reproductive fitness of both the nematode and bacterium and is therefore predictable in their integrated evolutionary life histories. When an IJ infects an insect, it gains entry into the insect hemocoel, into which it releases its Xenorhabdus symbiont (37–39). The hemocoel cavity is bathed in hemolymph, a relatively nutrient-rich fluid (40). IJ ingestion of insect hemolymph is always the first step leading to bacterial release into the insect hemocoel, similarly to the blood meals of a mosquito or a flea before the transmission of vector-borne pathogens. In Yersinia pestis, the temperature of and nutrients in mammalian blood during the flea feeding induce phenotypic variation and changes in bacterial virulence gene expression prior to introduction of the bacteria into the host itself (41–43). Similarly, X. nematophila exposure to hemolymph prior to release into the insect hemocoel may lead to preinduction of virulence genes, perhaps through sensing of its high concentrations of sugars (trehalose and glucose) (44–46).
FIG 2.
Series of Steinernema host intestinal environments encountered by Xenorhabdus. (A to D) Simplified intestinal structures of nematodes (not whole organisms) at different stages (IJ, adult and juvenile, and pre-IJ) are schematically represented, with Xenorhabdus bacteria indicated by green ovals. (A) Once an IJ nematode enters the insect hemocoel from the soil, X. nematophila bacteria (green ovals) are released from the widening intestinal lumen (IL) of the recovering IJ into insect hemolymph during infection. (B) Adult and juvenile nematodes in the insect cadaver are colonized by symbiotic bacteria at the anterior intestinal cecum (AIC), a region within the intestine immediately below the pharyngeal intestinal valve (PIV). (C) In a pre-IJ nematode, a few symbiotic bacterial cells are enclosed in pouches within the PIV. (D) In an IJ nematode with a closed intestine, symbiotic bacteria colonize the receptacle. Bacteria either are associated with the intravesicular structure (IVS) or move freely in the receptacle lumen. (E) X. nematophila bacteria expressing low levels of Lrp are more virulent toward insects. Blue curve, high-Lrp-expressing bacteria; red curve, low-Lrp-expressing bacteria. (F) X. nematophila bacteria expressing high levels of Lrp better support nematode reproduction in the insect cadaver. Blue curve, high-Lrp-expressing bacteria; red curve, low-Lrp-expressing bacteria. (G) X. nematophila bacteria expressing high levels of Lrp show a high colonization frequency in IJ nematodes. bb, basal bulb; IL, intestinal lumen; AIC, anterior intestinal cecum; PIV, pharyngeal intestinal valve; IVS, intravesicular structure.
The transition of Xenorhabdus from the aging IJ receptacle to the hemocoel of an insect represents a dramatic shift in selective environment. The hemocoel is relatively nutrient rich but is a primary site for host surveillance and induction of immune responses. Insect cellular immunity involves hemocytes that engulf bacteria and help clear bacteria through melanin production. Humoral immune responses include the production of antimicrobial peptides that can lyse bacteria (47). In the insect hemocoel, bacterial survival depends on rapidly countering immunity and killing the host, and, indeed, X. nematophila can suppress insect immunity and produces virulence factors that contribute to insect host death (26, 48). Preadaptive responses that prime Xenorhabdus expression of immunosuppressive activities and toxins while still within the IJ receptacle could provide a selective advantage.
FROM VIRULENCE TO FEEDING
A second predictable change in environment occurs upon death of the insect (arrow between panels A and B in Fig. 2). The insect cadaver serves as a nutrient source for bacterial growth and nematode reproduction, and Xenorhabdus bacteria are key players in the liberation of the nutrients (carbohydrates, lipids, and amino acids) contained within the insect biomass (26). Events that may precede and indicate impending insect death, and that therefore may serve as signals for preadaptive responses, are the release of nutrients from dying hemocytes (49), the growth of competing microorganisms (50), or changes in the insect intestinal barrier (39).
Xenorhabdus bacteria are not necessarily only living freely within the insect cadaver. Recent discoveries revealed that during the insect-degradation phase of the symbiotic life cycle, X. nematophila cells colonize the intestinal epithelium of all stages of the developing nematode host. In adult and juvenile stages of nematodes, multiple Xenorhabdus cells attach to the epithelial surface of the anterior intestinal cecum (AIC) (Fig. 2B) (51). The consequences of this colonization for either Xenorhabdus or Steinernema have not been elucidated, but these observations remind us that the insect cadaver does not represent a single homogenous environment but rather that different X. nematophila cells within the population may be encountering various challenges and various levels of nutritional content, depending on whether they are free-living in the liquefied insect cadaver, associated with insect tissues, attaching to the nematode AIC region, or passing through the nematode intestinal lumen while the nematode is actively digesting. Despite its complexity in the spatial distribution of nutrients, the local environments in the insect cadaver could play a role similar to those in the mammalian GI tract, driving phenotypic heterogeneity and preadaptive responses in symbiotic bacteria.
FROM THE SPENT CADAVER TO THE IJ
The depletion of nutrients and high population densities prompt Xenorhabdus colonization of the developing IJ receptacle. This process has not yet been observed within an individual nematode, but population-wide studies suggest that it, like the other stages of the Xenorhabdus life cycle, consists of a temporally ordered series of events. The pre-IJ is characterized by a collapsing intestinal tract, and during this stage, Xenorhabdus bacteria are no longer observed at the AIC; instead, individual cells colonize the pharyngeal intestinal valve (PIV), in a pocket formed by nematode tissues (Fig. 2C). As pre-IJ nematodes undergo further morphological changes and develop into a nonfeeding stage of IJ nematodes, the receptacle (intestinal pocket) is formed in a completely closed intestine, and individual bacterial cells are observed in this location, rather than at the pharyngeal intestinal valve (51) (Fig. 2D). During this IJ colonization initiation phase, a few Xenorhabdus bacterial cells localize in the newly formed receptacle. These subsequently grow very slowly into a clonal (or nearly clonal) population (52) that persists until they are introduced a new insect host. The serial events during the process of Xenorhabdus bacteria transitioning from residing in insect cadavers to associating with IJ nematodes features a predictable nutrient change of local environments, which could cause adaptive changes in gene expression. What signals occur within the insect cadaver that might trigger preadaptive modulation of gene expression optimal for colonization? The receptacle of Steinernema IJ nematodes contains a wheat-germ-agglutinin-reactive mucus-like substance, which perhaps is utilized by X. nematophila as a nutrient source within this environment (53). Preadaptive responses to this substance may be triggered by similar glycans expressed on the AIC surface of developing nematodes or by other sugars present in the insect cadaver. Other potential candidate signaling molecules could be derived from the insect cadaver. This idea is supported by evidence that nematodes grown in the insect better associate with bacterial symbionts than those reared in vitro on nutrient agar bacterial lawns (29, 30). This suggests that metabolites specifically derived from the insect tissues (via either nematode or bacterial metabolism) may be important for symbiont transmission and nutrient adaptation. Nematodes also may be a source of preadaptive signals for colonization. For instance, Steinernema spp. and other nematodes secrete a variety of ascarosides, signaling pheromones that can regulate social behaviors such as mating, development, and dispersal (54–56). In the nematode Caenorhabditis elegans, the accumulation of specific ascarosides at high population densities triggers juvenile nematodes to enter the dauer stage, a nonfeeding larval stage similar to the Steinernema IJ (55). In addition, particular ascaroside molecules produced in Steinernema IJs signal nematode dispersal behavior that leads to their emergence from insect cadavers into the soil (54). Since the timing of ascaroside accumulation may synchronize with or precede the process of bacterial colonization during IJ formation, they are excellent candidates for signals in preadaptive responses. Finally, molecules produced by Xenorhabdus bacteria may themselves be preadaptive signals. Xenorhabdus secondary metabolites are produced during the reproductive phase of the life cycle and can antagonize invading microbial species to protect the insect cadaver (50, 57). Changes in the local concentrations of these metabolites could be indicative of conditions that warrant IJ development and emergence.
As mentioned above, the insect cadaver is spatially heterogeneous and may present distinctive signals that promote specialized adaptive responses. Future research on the identification and quantification of metabolites that are (both spatially and temporally) derived from insect tissues, nematodes, and bacteria in the insect cadaver will help in elucidating the particular environmental changes taking place during the host-switching events and the mechanisms in bacterial (pre)adaptive responses.
PERSISTENCE IN THE IJ
Once a few bacterial cells colonize the IJ receptacle, they grow slowly into a population that persists in the host for periods lasting up to months until the nematode enters a new insect host (30, 52). The IJ receptacle has some spatial and temporal nutrient complexity that could help trigger adaptive gene expression as IJs age. The closed mouth and collapsed intestine in the nonfeeding IJs (28) make the receptacle a relatively closed environment, restricting nutrient exchange with the outside. The receptacle is the lumen between two anterior intestinal cells that are morphologically distinct from the rest of the intestinal cells (53). Also, in some Steinernema nematodes, Xenorhabdus symbionts are enclosed in a cellophane-like envelope membrane termed the vesicle (58). The receptacle (or the vesicle in those nematodes) contains an intravesicular structure (IVS) that consists of a cluster of anucleate particles surrounded by mucus-like material (Fig. 2D). This may indicate spatial distribution of nutrients, which could create heterogeneous local environments for the symbionts. Indeed, within the receptacle, Xenorhabdus bacteria can be observed attached to the IVS surface, tightly packed with other bacterial cells, or individually as unattached and free-floating in the receptacle space (53). Nutrient availability in the IJ receptacle is likely to vary temporally during IJ host persistence in the soil, and such changes could contribute to preadaptive responses to the following insect host environment. Although the IJ does not itself receive fresh influxes of nutrients, it may contribute various nutrients to its symbiont as it ages. Indeed, X. nematophila does have access to certain essential amino acids in the receptacle, since auxotrophs for those amino acids are able to colonize and grow there (32). Finally, IJs colonized by symbionts showed a reduced life span in comparison to aposymbiotic IJs (59), suggesting that bacteria may continuously exploit the host nutrients during the IJ aging process. During this nonfeeding stage, IJs store and utilize energy from lipid droplets that contain triglycerols, sterol esters, phospholipids, and proteins (60, 61). In Steinernema species, glycogen and trehalose reserves are continuously synthesized and significantly consumed during the IJ aging process (62, 63). It is not yet known if and how nematode transfer metabolites into the receptacle. Neither is it known if temporal changes in the nematode physiology (i.e., IJ maturation, movements, aging, etc.) affect the levels of nutrients provided to symbionts. Regardless, spatial complexity and temporal dynamics of nutrient composition in aging IJ receptacles may be signals that cause preadaption of colonizing Xenorhabdus to the next phase of the life cycle: the insect host.
PHENOTYPIC VARIATION IN X. NEMATOPHILA
The sequential environments encountered by X. nematophila described above set the stage for the evolution of adaptive regulatory pathways. One of these pathways is controlled by the Lrp global transcription factor. Lrp homologs are small (∼19-kDa) DNA binding proteins that are widely conserved among bacteria and archaea (64, 65). They are members of a larger conserved family of transcriptional regulators known as the feast/famine regulatory proteins. These regulators contribute to nutrient adaptation by sensing various environmental signals and eliciting an adaptive global change in gene expression (66). In X. nematophila, Lrp is a global regulator that controls numerous genes with predicted roles in host interactions, motility, nutrient adaptation, and the production of small molecules (67, 68). An lrp mutant has defects in each stage of X. nematophila host interactions: it is defective in immune suppression and killing of M. sexta insects, supporting reproduction of and colonizing nematodes, and transitioning between nutrient-limiting and nutrient-rich media (67, 69).
A more complicated role for Lrp in the X. nematophila life cycle was recently reported, based on the discovery that it controls a phenotypic variation phenomenon known as virulence modulation (VMO) (68, 70). This phenomenon is marked by the spontaneous and reversible switch between virulent and virulence-attenuated phenotypes (as assessed by the ability to kill Manduca sexta insects upon injection) (68, 70). A role for Lrp in VMO was initially suggested by the finding that Lrp protein levels differ from cell to cell in wild-type populations and that these levels correlate with immunosuppressive and virulence phenotypes after injection into insects. Surprisingly, this correlation is inverse: X. nematophila cells expressing high levels of Lrp exhibit attenuated immune suppression and virulence, whereas those expressing low levels of Lrp are immunosuppressive and virulent (68) (Fig. 2E). The link between Lrp levels and virulence phenotypes has been substantiated using X. nematophila cells engineered to constitutively express high and low levels of Lrp. These cells exhibit attenuated and virulent phenotypes, respectively (68). More-recent studies have explored the biological role of the virulence-attenuated high Lrp cell type, revealing that high-Lrp expressers are significantly better than low-Lrp expressers in supporting nematode reproduction and are slightly better at initiating colonization of IJs (Fig. 2F and G). In contrast, bacteria expressing constitutively low levels of Lrp are virulent but defective in supporting nematode reproduction and colonization (Fig. 2F and G) (68). Overall, these data suggest that an individual wild-type X. nematophila cell expresses one of two symbiotic gene expression profiles or switches between the two in a manner optimal for virulence or mutualism, depending on if it has low or high cellular levels of Lrp, respectively. Further, the high- or low-Lrp expresser state is heritable and reversible, suggesting that a phenotypic variation phenomenon controls the switch between high and low states (and therefore between mutualistic and virulent phenotypes) (68, 70, 71).
In bacterial phenotypic variation, transcriptional and translational noise among individual cells can be amplified by positive- or negative-feedback loops, causing the bacterial population to bifurcate into two inheritable and reversible phenotypes, a phenomenon called bistability (72). In X. nematophila, Lrp negatively regulates its own promoter, raising the possibility that Lrp bistable expression is controlled by a single autoregulatory negative-feedback loop (68). Alternatively, population-level bistability could also be caused by cell-level multistationarity, such as multimerization and phosphorylation of proteins, or by cooperative binding of DNA (72). Lrp has been implicated previously in regulation of phenotypic variation in other bacterial species through cellular multistationarity. For example, in uropathogenic E. coli (UPEC), through transcriptional regulation of the pap promoter, Lrp controls an on/off phenotypic switch of Pap fimbrial expression. UPEC Lrp binds to three of the six Lrp binding sites in the pap promoter; binding of proximal sites leads to the pap off state, while binding of distal sites results in the on state (73). Cocrystal structures of Lrp with pap promoter DNA reveal that DNA wraps around and binds to an octameric Lrp complex, reminiscent of the eukaryotic nucleosome-like complex (74–76). Therefore, Lrp multimerization and DNA topology are crucial for transcriptional regulation, providing a mechanistic rationale for how changes in Lrp concentrations could influence phenotypic variation of symbiotic phenotypes in X. nematophila. Whether VMO in X. nematophila is regulated by Lrp bistable or multistable expression remains to be tested.
These findings have led to the idea that the Lrp-dependent VMO switch gives rise to population heterogeneity with respect to host interactions and adaptive responses to environmental changes over the entire life history of X. nematophila. On the basis of current knowledge of the phenotypes of high- and low-Lrp-expressing cells, we have developed a working model of how Lrp-dependent phenotypic variation might contribute to adaptive prediction if it occurs in X. nematophila (Fig. 1C). Briefly, if adaptive prediction is occurring, each environment (Fig. 1C, top row, open symbols) either selects for induction of a phenotypic switch or selects for a particular cell type that expresses the Lrp-dependent gene expression profile (Fig. 1C, high- or low-Lrp expression; bottom row, closed symbols) that is adaptive for the next environment that will be encountered. This model of adaptive prediction awaits testing and is not mutually exclusive with the alternative, canonical model that the prevailing conditions elicit a gene expression response that is adaptive under that condition (not for a future condition). To begin to address these ideas, the next steps will be to investigate the temporal and spatial Lrp-dependent population heterogeneity over the span of the X. nematophila life cycle and to determine if this variability serves as a (pre)adaptive mechanism for bacteria transitioning from the nematode to the insect host.
CONCLUSIONS
Anticipatory behavior, or the ability to learn from history, has been long known in vertebrates (77) and has recently been investigated in microbes. During the symbiotic life cycle, microbes often are exposed to and must adapt to predictable or indeterminate environmental changes in a temporal and spatial manner, which could cause population heterogeneity and anticipatory behavior. Current experimental evidence of microbial anticipation in an animal host environment is limited to E. coli. Here we have discussed multiple animal-microbe symbiosis models that hint at the possibility of anticipatory behavior contributing to the fitness and success of the microbial symbiont partner. Testing this concept will rely on the establishment of the signals that characterize the spatial and temporal environments encountered by symbionts and on testing the abilities of these signals to elicit preadaptive responses. The symbiosis between Xenorhabdus bacteria and Steinernema nematodes provides a powerful system with which to approach this type of research, based on its predictability for both the bacterium and the investigator. As we discussed, there is considerable knowledge about the stages, microenvironments, and regulatory processes of the Xenorhabdus life cycle. Together, these provide a strong foundation for formulating testable hypotheses about the triggers and outputs of possible anticipatory behaviors in symbiosis.
REFERENCES
- 1.Wales AD, Woodward MJ, Pearson GR. 2005. Attaching-effacing bacteria in animals. J Comp Pathol 132:1–26. doi: 10.1016/j.jcpa.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs JL, Skophammer RG, Regus JU. 2011. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci U S A 108(Suppl):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas AE. 2010. The symbiotic habit. Princeton University Press, Princeton, New Jersey. [Google Scholar]
- 4.Chaston J, Goodrich-Blair H. 2010. Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol Rev 34:41–58. doi: 10.1111/j.1574-6976.2009.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acar M, Mettetal JT, van Oudenaarden A. 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 6.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 8.Tagkopoulos I, Liu Y-C, Tavazoie S. 2008. Predictive behavior within microbial genetic networks. Science 320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keim PS, Wagner DM. 2009. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nat Rev Microbiol 7:813–821. doi: 10.1038/nrmicro2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heath-Heckman EAC. 2016. The metronome of symbiosis: interactions between microbes and the host circadian clock. Integr Comp Biol 56:776–783. doi: 10.1093/icb/icw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Woude MW. 2011. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol 14:205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 13.Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun 73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog 6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsoumtsa LL, Torre C, Ghigo E. 2016. Circadian control of antibacterial immunity: findings from animal models. Front Cell Infect Microbiol 6:54. doi: 10.3389/fcimb.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Man K, Loudon A, Chawla A. 2016. Immunity around the clock. Science 354:999–1003. doi: 10.1126/science.aah4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyholm SV, McFall-Ngai MJ. 1998. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull 195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 20.Lamarcq LH, McFall-Ngai MJ. 1998. Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by Vibrio fischeri. Infect Immun 66:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath-Heckman EAC, Foster J, Apicella MA, Goldman WE, McFall-Ngai M. 2016. Environmental cues and symbiont microbe-associated molecular patterns function in concert to drive the daily remodelling of the crypt-cell brush border of the Euprymna scolopes light organ. Cell Microbiol 18:1642–1652. doi: 10.1111/cmi.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo MDF, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A 107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heath-Heckman EAC, McFall-Ngai MJ. 2011. The occurrence of chitin in the hemocytes of invertebrates. Zoology 114:191–198. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartzman JA, Koch E, Heath-Heckman EAC, Zhou L, Kremer N, McFall-Ngai MJ, Ruby EG. 2015. The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci U S A 112:556–571. doi: 10.1073/pnas.1418580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heath-Heckman EAC, Peyer SM, Whistler CA, Apicella MA, Goldman WE, Mcfall-ngai MJ. 2013. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio 4:e00167-13. doi: 10.3391/mbi.2013.4.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards GR, Goodrich-Blair H. 2009. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol 11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morran LT, Penley MJ, Byrd VS, Meyer AJ, O'Sullivan TS, Bashey F, Goodrich-Blair H, Lively CM. 2016. Nematode-bacteria mutualism: selection within the mutualism supersedes selection outside of the mutualism. Evolution 70:687–695. doi: 10.1111/evo.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird AF, Akhurst RJ. 1983. The nature of the intestinal vesicle in nematodes of the family steinernematidae. Int J Parasitol 13:599–606. doi: 10.1016/S0020-7519(83)80032-0. [DOI] [Google Scholar]
- 29.Flores-Lara Y, Renneckar D, Forst S, Goodrich-Blair H, Stock P. 2007. Influence of nematode age and culture conditions on morphological and physiological parameters in the bacterial vesicle of Steinernema carpocapsae (Nematoda: Steinernematidae). J Invertebr Pathol 95:110–118. doi: 10.1016/j.jip.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Goetsch M, Owen H, Goldman B, Forst S. 2006. Analysis of the PixA inclusion body protein of Xenorhabdus nematophila. J Bacteriol 188:2706–2710. doi: 10.1128/JB.188.7.2706-2710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jubelin G, Pagès S, Lanois A, Boyer M-H, Gaudriault S, Ferdy J-B, Givaudan A. 2011. Studies of the dynamic expression of the Xenorhabdus FliAZ regulon reveal atypical iron-dependent regulation of the flagellin and haemolysin genes during insect infection. Environ Microbiol 13:1271–1284. doi: 10.1111/j.1462-2920.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- 32.Martens EC, Russell FM, Goodrich-Blair H. 2005. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol Microbiol 58:28–45. doi: 10.1111/j.1365-2958.2005.04742.x. [DOI] [PubMed] [Google Scholar]
- 33.Ishibashi N, Kondo E. 1986. Steinernema feltiae (DD-136) and S. glaseri: persistence in soil and bark compost and their influence on native nematodes. J Nematol 18:310–316. [PMC free article] [PubMed] [Google Scholar]
- 34.Kung S-P, Gaugler R. 1991. Effects of soil temperature, moisture, and relative humidity on entomopathogenic nematode persistence. J Invertebr Pathol 57:242–249. doi: 10.1016/0022-2011(91)90123-8. [DOI] [Google Scholar]
- 35.Kung SP, Gaugler R, Kaya HK. 1990. Influence of soil pH and oxygen on persistence of Steinernema spp. J Nematol 22:440–445. [PMC free article] [PubMed] [Google Scholar]
- 36.Kung S-P, Gaugler R. 1990. Soil type and entomopathogenic nematode persistence. J Invertebr Pathol 55:401–406. doi: 10.1016/0022-2011(90)90084-J. [DOI] [Google Scholar]
- 37.Poinar GO, Thomas GM. 1967. The nature of Achromobacter nematophilus as an insect pathogen. J Invertebr Pathol 9:510–514. doi: 10.1016/0022-2011(67)90131-0. [DOI] [Google Scholar]
- 38.Snyder H, Stock SP, Kim S-KK, Flores-Lara Y, Forst S. 2007. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl Environ Microbiol 73:5338–5346. doi: 10.1128/AEM.02947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sicard M, Brugirard-ricaud K, Page S, Lanois A, Boemare NE, Brehe M, Givaudan A, Gpia L. 2004. Stages of infection during the tripartite interaction between Xenorhabdus nematophila, its nematode vector, and insect hosts. Appl Environ Microbiol 70:6473–6480. doi: 10.1128/AEM.70.11.6473-6480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phalaraksh C, Lenz EM, Lindon JC, Nicholson JK, Farrant RD, Reynolds SE, Wilson ID, Osborn D, Weeks JM. 1999. NMR spectroscopic studies on the haemolymph of the tobacco hornworm, Manduca sexta: assignment of 1H and 13C NMR spectra. Insect Biochem Mol Biol 29:795–805. doi: 10.1016/S0965-1748(99)00053-3. [DOI] [Google Scholar]
- 41.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 43.Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PCF, Hinnebusch BJ. 2013. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol 195:1920–1930. doi: 10.1128/JB.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodaki A, Bohovych IM, Enjalbert B, Young T, Odds FC, Gow NAR, Brown AJP. 2009. Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol Biol Cell 20:4845–4855. doi: 10.1091/mbc.E09-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalscheuer R, Koliwer-Brandl H. 23 May 2014 Genetics of mycobacterial trehalose metabolism. Microbiol Spectr doi: 10.1128/microbiolspec.MGM2-0002-2013. [DOI] [PubMed] [Google Scholar]
- 46.Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 47.Casanova-Torres ÁM, Goodrich-Blair H. 2013. Immune signaling and antimicrobial peptide expression in Lepidoptera. Insects 4:320–338. doi: 10.3390/insects4030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herbert EE, Goodrich-Blair H. 2007. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol 5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- 49.Cowles KN, Goodrich-Blair H. 2005. Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell Microbiol 7:209–219. doi: 10.1111/j.1462-5822.2004.00448.x. [DOI] [PubMed] [Google Scholar]
- 50.Singh S, Orr D, Divinagracia E, McGraw J, Dorff K, Forst S. 2015. Role of secondary metabolites in establishment of the mutualistic partnership between Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl Environ Microbiol 81:754–764. doi: 10.1128/AEM.02650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaston JM, Murfin KE, Heath-Heckman EA, Goodrich-Blair H. 2013. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cell Microbiol 15:1545–1559. doi: 10.1111/cmi.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martens EC, Heungens K, Goodrich-Blair H. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol 185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martens EC, Goodrich-Blair H. 2005. The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cell Microbiol 7:1723–1735. doi: 10.1111/j.1462-5822.2005.00585.x. [DOI] [PubMed] [Google Scholar]
- 54.Kaplan F, Alborn HT, von Reuss SH, Ajredini R, Ali JG, Akyazi F, Stelinski LL, Edison AS, Schroeder FC, Teal PE. 2012. Interspecific nematode signals regulate dispersal behavior. PLoS One 7:e38735. doi: 10.1371/journal.pone.0038735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ludewig AH, Schroeder FC. 18 January 2013 Ascaroside signaling in C. elegans. In The C. elegans Research Community, WormBook (ed), WormBook. doi: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, Schroeder FC, Sternberg PW. 2012. Ascaroside signaling is widely conserved among nematodes. Curr Biol 22:772–780. doi: 10.1016/j.cub.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park D, Forst S. 2006. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol Microbiol 61:1397–1412. doi: 10.1111/j.1365-2958.2006.05320.x. [DOI] [PubMed] [Google Scholar]
- 58.Sugar DR, Murfin KE, Chaston JM, Andersen AW, Richards GR, DeLéon L, Baum JA, Clinton WP, Forst S, Goldman BS, Krasomil-Osterfeld KC, Slater S, Stock SP, Goodrich-Blair H. 2012. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ Microbiol 14:924–939. doi: 10.1111/j.1462-2920.2011.02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitani DK, Kaya HK, Goodrich-Blair H. 2004. Comparative study of the entomopathogenic nematode, Steinernema carpocapsae, reared on mutant and wild-type Xenorhabdus nematophila. Biol Control 29:382–391. doi: 10.1016/j.biocontrol.2003.07.005. [DOI] [Google Scholar]
- 60.Patel MN, Stolinski M, Wright DJ. 1997. Neutral lipids and the assessment of infectivity in entomopathogenic nematodes: observations on four Steinernema species. Parasitology 114:489–496. doi: 10.1017/S0031182096008748. [DOI] [PubMed] [Google Scholar]
- 61.Bartz R, Li W-H, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RGW, Liu P, Chapman KD. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res 48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 62.Patel MN, Wright DJ. 1997. Glycogen: its importance in the infectivity of aged juveniles of Steinernema carpocapsae. Parasitology 114:591–596. doi: 10.1017/S0031182096008748. [DOI] [PubMed] [Google Scholar]
- 63.Qiu L, Lacey MJ, Bedding RA. 2000. Using deuterium as an isotopic tracer to study the energy metabolism of infective juveniles of Steinernema carpocapsae under aerobic conditions. Comp Biochem Physiol Part B Biochem Mol Biol 127:279–288. doi: 10.1016/S0305-0491(00)00253-4. [DOI] [PubMed] [Google Scholar]
- 64.Platko JV, Calvo JM. 1993. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J Bacteriol 175:1110–1117. doi: 10.1128/jb.175.4.1110-1117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci U S A 99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yokoyama K, Ishijima SA, Clowney L, Koike H, Aramaki H, Tanaka C, Makino K, Suzuki M. 2006. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev 30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 67.Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. 2007. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol 9:1311–1323. doi: 10.1111/j.1462-5822.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- 68.Hussa EA, Casanova-Torres Á, Goodrich-Blair MH. 2015. The global transcription factor Lrp controls virulence modulation in Xenorhabdus nematophila. J Bacteriol 197:3015–3025. doi: 10.1128/JB.00272-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heungens K, Cowles CE, Goodrich-Blair H. 2002. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol Microbiol 45:1337–1353. doi: 10.1046/j.1365-2958.2002.03100.x. [DOI] [PubMed] [Google Scholar]
- 70.Park Y, Herbert EE, Cowles CE, Cowles KN, Menard ML, Orchard SS, Goodrich-Blair H. 2007. Clonal variation in Xenorhabdus nematophila virulence and suppression of Manduca sexta immunity. Cell Microbiol 9:645–656. doi: 10.1111/j.1462-5822.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- 71.Cao M, Patel T, Goodrich-Blair H, Hussa EA. 7 April 2017 High levels of Xenorhabdus nematophila transcription factor Lrp promote mutualism with Steinernema carpocapsae nematode hosts. Appl Environ Microbiol doi: 10.1128/AEM.00276-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 73.van der Woude M, Braaten B, Low D. 1996. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol 4:5–9. doi: 10.1016/0966-842X(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 74.D'Ari R, Lin RT, Newman EB. 1993. The leucine-responsive regulatory protein: more than a regulator? Trends Biochem Sci 18:260–263. doi: 10.1016/0968-0004(93)90177-O. [DOI] [PubMed] [Google Scholar]
- 75.Beloin C, Jeusset J, Revet B, Mirambeau G, Le Hégarat F, Le Cam E. 2003. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J Biol Chem 278:5333–5342. doi: 10.1074/jbc.M207489200. [DOI] [PubMed] [Google Scholar]
- 76.de los Rios S, Perona JJ. 2007. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J Mol Biol 366:1589–1602. doi: 10.1016/j.jmb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pavlov IP. 1927. Conditioned reflexes: an investigation of the physiological activity of the cerebral cortex. Oxford University Press, Oxford, England. [DOI] [PMC free article] [PubMed] [Google Scholar]