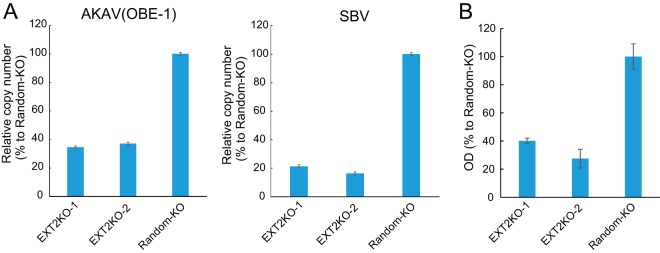
FIG 3.
AKAV and SBV binding assays in HSPG-KO cells. (A) Real-time RT-PCR for the quantification of cell surface-attached viruses. AKAV(OBE-1) or SBV was incubated with HSPG-KO cells at 4°C. After a washing step, total RNAs were extracted. AKAV or SBV S RNAs were quantified by one-step real-time RT-PCR. For relative quantification, standard curves of AKAV or SBV S RNA and GAPDH were prepared by serial dilution of a mixture of total RNA from uninfected HmLu-1 cells and RNA extracted from AKAV(OBE-1) or SBV-containing supernatants. Results are expressed as the percentages relative to the levels in random-KO cells. The results are representative of three different experiments. (B) Sandwich ELISA for the detection of N proteins of AKAV attached to cell surfaces. AKAV(OBE-1) was inoculated onto HSPG-KO or random-KO HmLu-1 cells and left for 1 h at 4°C. After a washing step, the cells were lysed, and the lysates were added to the anti-AKAV N monoclonal antibody (5E8)-coated wells of 96-well ELISA plates (Maxisorp, Nunc), followed by incubation with biotinylated anti-AKAV mouse polyclonal antibody. Subsequently, the wells were incubated with avidin-biotinylated horseradish peroxidase (HRP) complex (Vectastain ABC kit; Vector Laboratories). A 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution was used for detection, and optical density values were measured. Results are expressed as percentages relative to the number of positive random-KO cells. The results are representative of three independent experiments.