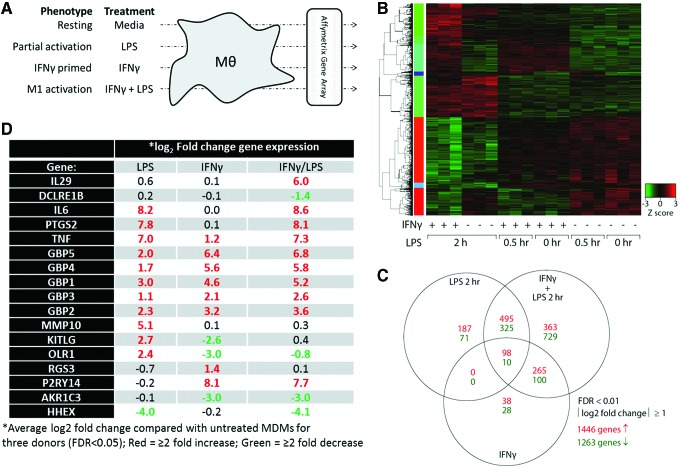
FIG. 1.
Gene array transcriptome analysis of macrophage activation states. (A) MDM were cultured for 7 days. On day 6, some samples were primed with 10 ng/mL IFNγ for 18 h. On day 7, MDM were left untreated or activated with 10 ng/mL LPS for 0.5 or 2 h. Total RNA was used to perform Affymetrix U133 Plus 2.0 gene arrays. (B) Normalized data were analyzed by hierarchical clustering to determine gene expression clusters. (C) The unique or shared expression of genes representing partial activation (2 h LPS), IFNγ priming, or classical M1 activation (IFNγ + 2 h LPS) that showed at least a twofold change in expression is represented by a Venn diagram. (D) Genes that uniquely identify different activation states were selected for further analyses. Experiment was conducted in three donors. MDM, monocyte-derived macrophages. Color images available online at www.liebertpub.com/aid