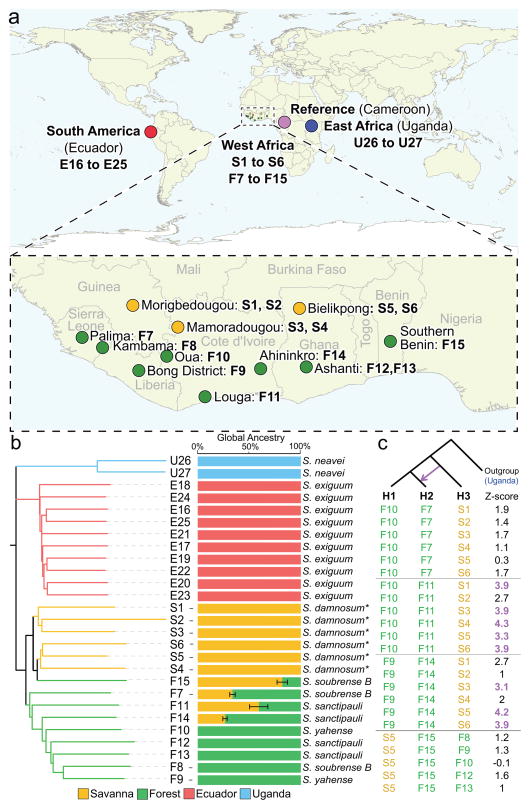
Figure 1.
The geographical and genomic relationship between O. volvulus isolates. (a) Twenty- seven individual isolates were collected from Ecuador, Uganda and 11 sites in West Africa (3 savanna and 8 forest sites). (b) Clustering of isolates based on single nucleotide polymorphisms in the three nuclear autosomes, and admixture analysis was used to assess patterns of global ancestry among samples. Error bars represent standard errors estimated using bootstrapping. Local Simulium vector species are presented for each isolate. * or S. sirbanum (c) Gene flow between allopatric populations (Patterson’s D test). Isolate F15 was treated as a savanna sample, and showed no significant gene flow from any forest isolate. Numbers colored in purple were significant according to the test (z-score > 3).