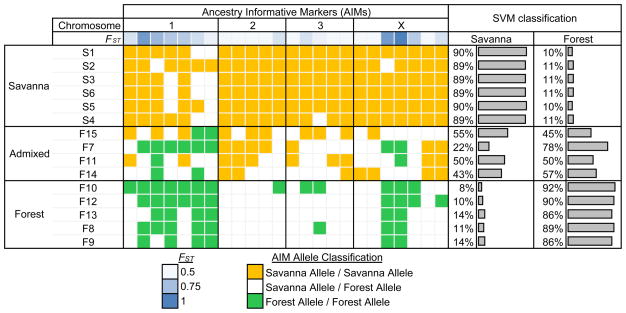
Figure 3.
Ancestry-informative markers (AIMs) that exhibit substantially different allele frequencies between the West African forest and savanna populations were used to assign individual isolates to source populations and identify genetic admixture. These 24 markers were selected based on informativeness (i.e., high FST values) and genome-wide distribution (Supplementary Table 7). Prediction of the individuals’ population membership based on Support Vector Machine (SVM) shows that the AIMs provide the discriminatory power to assign individuals to the correct origin and identify admixture.