Figure 3.
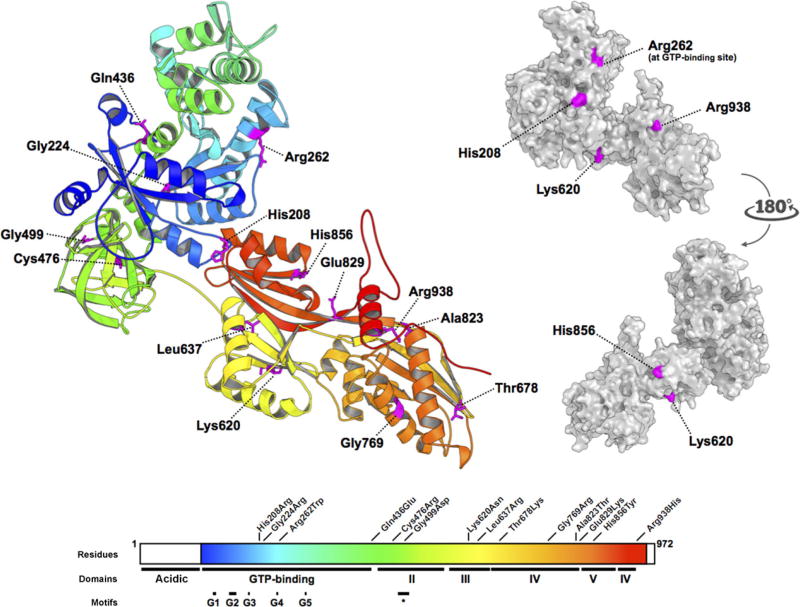
All EFTUD2 missense substitutions described to date. (Based on Fabrizio et al., 1997 and Lines et al., 2012). Residues 114–957 of EFTUD2 were modelled on the crystal structure of S. cerevisiae ribosomal elongation factor 2 (eEF2) (PDB: 1N0U). Missense substitutions are distributed throughout all domains of the protein. Conserved motifs identified by Fabrizio et al. (1997), including the GTP-binding domains G1 through G5, and the short conserved domain II motif (∗), are shown. The side chains of His208, Arg262, Lys620, and His856, and Arg938, all basic residues, are predicted to be surface-forming, whereas the sidechains of Gln436, Leu637, Thr678, and Glu829, Gly224, Cys476, Gly499, Gly769, and Ala823 are interior to the model.